Identification of Therapeutic Targets for Medulloblastoma by Tissue-Specific Genome-Scale Metabolic Model
Abstract
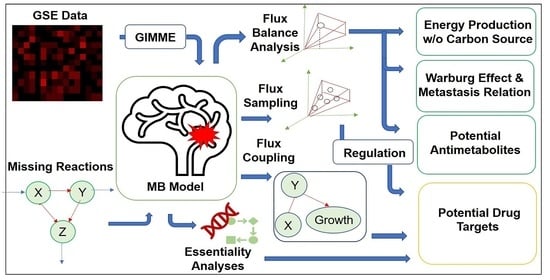
:1. Introduction
2. Results
2.1. Energy Production and Growth Rate without Glucose and Glutamine Sources
2.2. The Relationship between Metastasis and the Warburg Effect
2.3. Flux Sampling and Regulation of Reactions
2.4. Drug Targets for Medulloblastoma
2.4.1. Targeting Biomass-Coupled Reactions to Inhibit Tumor Growth
2.4.2. Genes and Gene Products as Targets for Suppressing Tumor Growth: Essentiality Analyses
2.4.3. Targeting Common Essential Genes in Biomass, Lactate, and Energy Productions
2.4.4. Antimetabolites Competing with Natural Metabolites
3. Discussion
4. Materials and Methods
4.1. Reconstruction
4.1.1. Transcriptomic Data Integration
4.1.2. Constraints
4.2. Operation
Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Girardi, F.; Allemani, C.; Coleman, M.P. Worldwide Trends in Survival from Common Childhood Brain Tumors: A Systematic Review. J. Glob. Oncol. 2019, 5, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Northcott, P.A.; Robinson, G.W.; Kratz, C.P.; Mabbott, D.J.; Pomeroy, S.L.; Clifford, S.C.; Rutkowski, S.; Ellison, D.W.; Malkin, D.; Taylor, M.D.; et al. Medulloblastoma. Nat. Rev. Dis. Prim. 2019, 5, 11. [Google Scholar] [CrossRef]
- Wang, J.; Garancher, A.; Ramaswamy, V.; Wechsler-Reya, R.J. Medulloblastoma: From molecular subgroups to molecular targeted therapies. Annu. Rev. Neurosci. 2018, 41, 207–232. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Kumar, V.; McGuire, T.; Coulter, D.W.; Sharp, J.G.; Mahato, R.I. Challenges and Recent Advances in Medulloblastoma Therapy. Trends Pharmacol. Sci. 2017, 38, 1061–1084. [Google Scholar] [CrossRef] [PubMed]
- Palmer, S.L.; Armstrong, C.; Onar-Thomas, A.; Wu, S.; Wallace, D.; Bonner, M.J. Processing speed, attention, and working memory after treatment for medulloblastoma: An international, prospective, and longitudinal study. J. Clin. Oncol. 2013, 31, 3494–3500. [Google Scholar] [CrossRef] [Green Version]
- Riggs, L.; Bouffet, E.; Laughlin, S.; Laperriere, N.; Liu, F.; Skocic, J.; Scantlebury, N.; Wang, F.; Schoenhoff, N.J.; Strother, D.; et al. Changes to memory structures in children treated for posterior fossa tumors. J. Int. Neuropsychol. Soc. 2014, 20, 168–180. [Google Scholar] [CrossRef] [Green Version]
- Folger, O.; Jerby, L.; Frezza, C.; Gottlieb, E.; Ruppin, E.; Shlomi, T. Predicting selective drug targets in cancer through metabolic networks. Mol. Syst. Biol. 2011, 7, 501. [Google Scholar] [CrossRef]
- Larsson, I.; Uhlén, M.; Zhang, C.; Mardinoglu, A. Genome-Scale Metabolic Modeling of Glioblastoma Reveals Promising Targets for Drug Development. Front. Genet. 2020, 11, 381. [Google Scholar] [CrossRef]
- Özcan, E.; Çakir, T. Reconstructed metabolic network models predict flux-level metabolic reprogramming in glioblastoma. Front. Neurosci. 2016, 10, 156. [Google Scholar] [CrossRef] [Green Version]
- Tech, K.; Gershon, T.R. Energy metabolism in neurodevelopment and medulloblastoma. Transl. Pediatr. 2015, 4, 12–129. [Google Scholar] [CrossRef]
- Warburg, O. The Metabolism of Carcinoma Cells. J. Cancer Res. 1925, 9, 148–163. [Google Scholar] [CrossRef] [Green Version]
- Munford, H. Investigating Medulloblastoma metabolism for better diagnosis and treatment. Ph.D. Thesis, University of Birmingham, Birmingham, UK, 2019. [Google Scholar]
- Bensaad, K.; Tsuruta, A.; Selak, M.A.; Vidal, M.N.C.; Nakano, K.; Bartrons, R.; Gottlieb, E.; Vousden, K.H. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell 2006, 126, 107–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niklison-Chirou, M.V.; Erngren, I.; Engskog, M.; Haglöf, J.; Picard, D.; Remke, M.; McPolin, P.H.R.; Selby, M.; Williamson, D.; Clifford, S.C.; et al. TAp73 is a marker of glutamine addiction in medulloblastoma. Genes Dev. 2017, 31, 1738–1753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sertbaş, M.; Ülgen, K.; Çakir, T. Systematic analysis of transcription-level effects of neurodegenerative diseases on human brain metabolism by a newly reconstructed brain-specific metabolic network. FEBS Open Bio. 2014, 4, 542–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çakir, T.; Alsan, S.; Saybaşili, H.; Akin, A.; Ülgen, K.Ö. Reconstruction and flux analysis of coupling between metabolic pathways of astrocytes and neurons: Application to cerebral hypoxia. Theor. Biol. Med. Model. 2007, 4, 48. [Google Scholar] [CrossRef] [Green Version]
- Gershon, T.R.; Crowther, A.J.; Tikunov, A.; Garcia, I.; Annis, R.; Yuan, H.; Miller, C.R.; Macdonald, J.; Olson, J.; Deshmukh, M. Hexokinase-2-mediated aerobic glycolysis is integral to cerebellar neurogenesis and pathogenesis of medulloblastoma. Cancer Metab. 2013, 1, 2. [Google Scholar] [CrossRef] [Green Version]
- Villa, E.; Ali, E.S.; Sahu, U.; Ben-Sahra, I. Cancer cells tune the signaling pathways to empower de novo synthesis of nucleotides. Cancers 2019, 11, 688. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Sánchez, R.; Rodríguez-Enríquez, S.; Saavedra, E.; Marín-Hernández, A.; Gallardo-Pérez, J.C. The bioenergetics of cancer: Is glycolysis the main ATP supplier in all tumor cells. BioFactors 2009, 35, 209–225. [Google Scholar] [CrossRef]
- Bennett, C.D.; Kohe, S.E.; Gill, S.K.; Davies, N.P.; Wilson, M.; Storer, L.C.D.; Ritzmann, T.; Paine, S.M.L.; Scott, I.S.; Nicklaus-Wollenteit, I.; et al. Tissue metabolite profiles for the characterisation of paediatric cerebellar tumours. Sci. Rep. 2018, 8, 11992. [Google Scholar] [CrossRef]
- Davies, N.P.; Wilson, M.; Harris, L.M.; Natarajan, K.; Lateef, S.; MacPherson, L.; Sgouros, S.; Grundy, R.G.; Arvanitis, T.N.; Peet, A.C. Identification and characterisation of childhood cerebellar tumours by in vivo proton MRS. NMR Biomed. 2008, 21, 908–918. [Google Scholar] [CrossRef]
- Kool, M.; Koster, J.; Bunt, J.; Hasselt, N.E.; Lakeman, A.; van Sluis, P.; Troost, D.; Meeteren, N.; Caron, H.N.; Cloos, J.; et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLoS ONE 2008, 3, 3088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yizhak, K.; Ruppin, E.; le Dévédec, S.E.; Rogkoti, V.M.; van de Water, B.; Baenke, F. A computational study of the Warburg effect identifies metabolic targets inhibiting cancer migration. Mol. Syst. Biol. 2014, 10, 744. [Google Scholar] [CrossRef] [PubMed]
- Bordel, S.; Agren, R.; Nielsen, J. Sampling the solution space in genome-scale metabolic networks reveals transcriptional regulation in key enzymes. PLoS Comput. Biol. 2010, 6, 16. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhou, T.; Li, C.; Dai, Z.; Che, D.; Yao, Y.; Li, L.; Ma, J.; Yang, X.; Gao, G. High metastaticgastric and breast cancer cells consume oleic acid in an AMPK dependent manner. PLoS ONE 2014, 9, 97330. [Google Scholar] [CrossRef]
- Natali, F.; Siculella, L.; Salvati, S.; Gnoni, G.V. Oleic acid is a potent inhibitor of fatty acid and cholesterol synthesis in C6 glioma cells. J. Lipid Res. 2007, 48, 1966–1975. [Google Scholar] [CrossRef] [Green Version]
- Anna, I.; Bartosz, P.; Lech, P.; Halina, A. Novel strategies of Raman imaging for brain tumor research. Oncotarget 2017, 8, 85290–85310. [Google Scholar] [CrossRef] [Green Version]
- Buford, T. Macronutrient intake for physical activity. In Nutritional Supplements in Sports and Exercise; Humana Press: Totowa, NJ, USA, 2008; pp. 95–119. [Google Scholar] [CrossRef]
- Rodriguez, G.V.; Abrahamsson, A.; Turkina, M.V.; Dabrosin, C. Lysine in Combination with Estradiol Promote Dissemination of Estrogen Receptor Positive Breast Cancer via Upregulation of U2AF1 and RPN2 Proteins. Front. Oncol. 2020, 10, 2650. [Google Scholar] [CrossRef]
- Kang, J.S. Dietary restriction of amino acids for Cancer therapy. Nutr. Metab. 2020, 17, 20. [Google Scholar] [CrossRef] [Green Version]
- Cormerais, Y.; Pagnuzzi-Boncompagni, M.; Schrötter, S.; Giuliano, S.; Tambutté, E.; Endou, H.; Wempe, M.F.; Pagès, G.; Pouysségur, J.; Picco, V. Inhibition of the amino-acid transporter LAT1 demonstrates anti-neoplastic activity in medulloblastoma. J. Cell. Mol. Med. 2019, 23, 2711–2718. [Google Scholar] [CrossRef]
- Panosyan, E.H.; Wang, Y.; Xia, P.; Lee, W.N.P.; Pak, Y.; Laks, D.R.; Lin, H.J.; Moore, T.B.; Cloughesy, T.F.; Kornblum, H.I.; et al. Asparagine depletion potentiates the cytotoxic effect of chemotherapy against brain tumors. Mol. Cancer Res. 2014, 12, 694–702. [Google Scholar] [CrossRef] [Green Version]
- Nielson, J.R.; Rutter, J.P. Lipid-mediated signals that regulate mitochondrial biology. J. Biol. Chem. 2018, 293, 7517–7521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blunsom, N.J.; Cockcroft, S. CDP-Diacylglycerol Synthases (CDS): Gateway to Phosphatidylinositol and Cardiolipin Synthesis. Front. Cell Dev. Biol. 2020, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Xu, Z.; Duan, H.; Ji, H.; Zhen, Z.; Li, B.; Wang, H.; Tang, H.; Zhou, J.; Guo, T.; et al. Tumor-associated macrophage interleukin-β promotes glycerol-3-phosphate dehydrogenase activation, glycolysis and tumorigenesis in glioma cells. Cancer Sci. 2020, 111, 1979–1990. [Google Scholar] [CrossRef]
- Singh, G. Mitochondrial FAD-linked Glycerol-3-phosphate Dehydrogenase: A Target for Cancer Therapeutics. Pharmaceuticals 2014, 7, 192–206. [Google Scholar] [CrossRef] [Green Version]
- Chang, F.; Li, R.; Noon, K.; Gage, D.; Ladisch, S. Human medulloblastoma gangliosides. Glycobiology 1997, 7, 523–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimoto, Y.; Izumoto, S.; Suzuki, T.; Kinoshita, M.; Kagawa, N.; Wada, K.; Hashimoto, N.; Maruno, M.; Nakatsuji, Y.; Yoshimine, T. Ganglioside GM3 inhibits proliferation and invasion of glioma. J. Neurooncol. 2005, 71, 99–106. [Google Scholar] [CrossRef]
- Gu, Y.; Zhang, J.; Mi, W.; Yang, J.; Han, F.; Lu, X.; Yu, W. Silencing of GM3 synthase suppresses lung metastasis of murine breast cancer cells. Breast Cancer Res. 2008, 10, R1. [Google Scholar] [CrossRef] [Green Version]
- Hooper, C.M.; Hawes, S.M.; Kees, U.R.; Gottardo, N.G.; Dallas, P.B. Gene Expression Analyses of the Spatio-Temporal Relationships of Human Medulloblastoma Subgroups during Early Human Neurogenesis. PLoS ONE 2014, 9, 112909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schömel, N.; Geisslinger, G.; Wegner, M.S. Influence of glycosphingolipids on cancer cell energy metabolism. Prog. Lipid Res. 2020, 79, 101050. [Google Scholar] [CrossRef]
- Ermini, L.; Morganti, E.; Post, A.; Yeganeh, B.; Caniggia, I.; Leadley, M.; Faria, C.C.; Rutka, J.T.; Post, M. Imaging mass spectrometry identifies prognostic ganglioside species in rodent intracranial transplants of glioma and medulloblastoma. PLoS ONE 2017, 12, 176254. [Google Scholar] [CrossRef] [Green Version]
- Longee, D.C.; Wikstrand, C.J.; Mnsson, J.E.; He, X.; Fuller, G.N.; Bigner, S.H.; Fredman, P.; Svennerholm, L.; Bigner, D.D. Disialoganglioside GD2 in human neuroectodermal tumor cell lines and gliomas. Acta Neuropathol. 1991, 82, 45–54. [Google Scholar] [CrossRef]
- Balis, F.M.; Busch, C.M.; Desai, A.V.; Hibbitts, E.; Naranjo, A.; Bagatell, R.; Irwin, M.; Fox, E. The ganglioside G D2 as a circulating tumor biomarker for neuroblastoma. Pediatr. Blood Cancer 2020, 67, e28031. [Google Scholar] [CrossRef] [PubMed]
- Marie, S.K.N.; Shinjo, S.M.O. Metabolism and Brain Cancer. Clinics 2011, 66, 33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ventura, R.; Mordec, K.; Waszczuk, J.; Wang, Z.; Lai, J.; Fridlib, M.; Buckley, D.; Kemble, G.; Heuer, T.S. Inhibition of de novo Palmitate Synthesis by Fatty Acid Synthase Induces Apoptosis in Tumor Cells by Remodeling Cell Membranes, Inhibiting Signaling Pathways, and Reprogramming Gene Expression. EBioMedicine 2015, 2, 808–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martirosian, V.; Deshpande, K.; Zhou, H.; Shen, K.; Smith, K.; Northcott, P.; Lin, M.; Stepanosyan, V.; Das, D.; Remsik, J.; et al. Medulloblastoma uses GABA transaminase to survive in the cerebrospinal fluid microenvironment and promote leptomeningeal dissemination. Cell Rep. 2021, 35, 109302. [Google Scholar] [CrossRef]
- Jiang, S.; Yan, W. Succinate in the cancer-immune cycle. Cancer Lett. 2017, 390, 45–47. [Google Scholar] [CrossRef]
- Hawkins, C.C.; Ali, T.; Ramanadham, S.; Hjelmeland, A.B. Sphingolipid Metabolism in Glioblastoma and Metastatic Brain Tumors: A Review of Sphingomyelinases and Sphingosine-1-Phosphate. Biomolecules 2020, 10, 1357. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer 2017, 18, 33–50. [Google Scholar] [CrossRef] [Green Version]
- Lahiri, S.; Futerman, A.H. The metabolism and function of sphingolipids and glycosphingolipids. Cell. Mol. Life Sci. 2007, 64, 2270–2284. [Google Scholar] [CrossRef] [PubMed]
- Feltrin, S.; Ravera, F.; Traversone, N.; Ferrando, L.; Bedognetti, D.; Ballestrero, A.; Zoppoli, G. Sterol synthesis pathway inhibition as a target for cancer treatment. Cancer Lett. 2020, 493, 19–30. [Google Scholar] [CrossRef]
- Larner, J.; Jane, J.; Laws, E.; Packer, R.; Myers, C.; Shaffrey, M. A phase I-II trial of lovastatin for anaplastic astrocytoma and glioblastoma multiforme. Am. J. Clin. Oncol. 1998, 21, 579–583. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, A.M.; Aboul-Soud, M.A.M.; Han, J.; Al-Sheikh, Y.A.; Al-Abd, A.M.; El-Shemy, H.A. Transcriptional profiling of breast cancer cells in response to mevinolin: Evidence of cell cycle arrest, DNA degradation and apoptosis. Int. J. Oncol. 2016, 48, 1886–1894. [Google Scholar] [CrossRef] [Green Version]
- Macaulay, R.J.B.; Wang, W.; Dimitroulakos, J.; Becker, L.E.; Yeger, H. Lovastatin-induced apoptosis of human medulloblastoma cell lines in vitro. J. Neurooncol. 1999, 42, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Sheikholeslami, K.; Sher, A.A.; Lockman, S.; Kroft, D.; Ganjibakhsh, M.; Nejati-Koshki, K.; Shojaei, S.; Ghavami, S.; Rastegar, M. Simvastatin Induces Apoptosis in Medulloblastoma Brain Tumor Cells via Mevalonate Cascade Prenylation Substrates. Cancers 2019, 11, 994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Shrivastava, S.; Hassanali, M.; Stothard, P.; Chang, Z.; Woolsey, J. DrugBank: A comprehensive resource for in silico drug discovery and exploration. Nucleic Acids Res. 2006, 34, D668–D672. [Google Scholar] [CrossRef] [PubMed]
- Benakanakere, I.; Johnson, T.; Sleightholm, R.; Villeda, V.; Arya, M.; Bobba, R.; Freter, C.; Huang, C. Targeting cholesterol synthesis increases chemoimmuno-sensitivity in chronic lymphocytic leukemia cells. Exp. Hematol. Oncol. 2014, 3, 24. [Google Scholar] [CrossRef] [Green Version]
- Grube, S.; Dünisch, P.; Freitag, D.; Klausnitzer, M.; Sakr, Y.; Walter, J.; Kalff, R.; Ewald, C. Overexpression of fatty acid synthase in human gliomas correlates with the WHO tumor grade and inhibition with Orlistat reduces cell viability and triggers apoptosis. J. Neurooncol. 2014, 118, 277–287. [Google Scholar] [CrossRef]
- Ohno, T.; Awaya, J.; Kesado, T.; Nomura, S.; Omura, S. Mechanism of Action of CM-55, a Synthetic Analogue of the Antilipogenic Antibiotic Cerulenin. Antimicrob. Agents Chemother. 1974, 6, 387. [Google Scholar] [CrossRef] [Green Version]
- Slade, R.F.; Hunt, D.A.; Pochet, M.M.; Venema, V.J.; Hennigar, R.A. Characterization and inhibition of fatty acid synthase in pediatric tumor cell lines. Anticancer Res. 2003, 23, 1235–1243. [Google Scholar]
- Ceci, C.; Lacal, P.M.; Tentori, L.; de Martino, M.G.; Miano, R.; Graziani, G. Experimental Evidence of the Antitumor, Antimetastatic and Antiangiogenic Activity of Ellagic Acid. Nutrients 2018, 10, 1756. [Google Scholar] [CrossRef]
- Kahn, S.A.; Wang, X.; Nitta, R.T.; Gholamin, S.; Theruvath, J.; Hutter, G.; Azad, T.D.; Wadi, L.; Bolin, S.; Ramaswamy, V.; et al. Notch1 regulates the initiation of metastasis and self-renewal of Group 3 medulloblastoma. Nat. Commun. 2018, 9, 4121. [Google Scholar] [CrossRef] [Green Version]
- Peters, G.J. Novel developments in the use of antimetabolites. Nucleosides Nucleotides Nucleic Acids 2014, 33, 358–374. [Google Scholar] [CrossRef] [PubMed]
- Röhrig, F.; Schulze, A. The multifaceted roles of fatty acid synthesis in cancer. Nat. Rev. Cancer 2016, 16, 732–749. [Google Scholar] [CrossRef] [PubMed]
- Mullen, P.J.; Yu, R.; Longo, J.; Archer, M.C.; Penn, L.Z. The interplay between cell signalling and the mevalonate pathway in cancer. Nat. Rev. Cancer 2016, 16, 718–731. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Wang, N.N.; Yao, Z.J.; Zhang, L.; Cheng, Y.; Ouyang, D.; Lu, A.-P.; Cao, D.-S. Admetlab: A platform for systematic ADMET evaluation based on a comprehensively collected ADMET database. J. Cheminform. 2018, 10, 29. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Son, H.S.; Cui, Y.H.; Youn, B.H.; Son, B.; Kaushik, N.K.; Uddin, N.; Lee, J.-S.; Song, J.-Y.; Kaushik, N.; et al. Phytosphingosine exhibits an anti-epithelial-mesenchymal transition function by the inhibition of EGFR signaling in human breast cancer cells. Oncotarget 2017, 8, 77794–77808. [Google Scholar] [CrossRef] [Green Version]
- Nagahara, Y.; Shinomiya, T.; Kuroda, S.; Kaneko, N.; Nishio, R.; Ikekita, M. Phytosphingosine induced mitochondria-involved apoptosis. Cancer Sci. 2005, 96, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Park, M.T.; Choi, J.A.; Kim, M.J.; Um, H.D.; Bae, S.; Kang, C.M.; Cho, C.-K.; Kang, S.; Chung, H.Y.; Lee, Y.-S.; et al. Suppression of Extracellular Signal-related Kinase and Activation of p38 MAPK Are Two Critical Events Leading to Caspase-8- and Mitochondria-mediated Cell Death in Phytosphingosine-treated Human Cancer Cells. J. Biol. Chem. 2003, 278, 50624–50634. [Google Scholar] [CrossRef] [Green Version]
- Eun, H.A.; Chang, C.C.; Schroeder, J.J. Evaluation of sphinganine and sphingosine as human breast cancer chemotherapeutic and chemopreventive agents. Exp. Biol. Med. 2006, 231, 1664–1672. [Google Scholar] [CrossRef]
- Ryland, L.K.; Fox, T.E.; Liu, X.; Loughran, T.P.; Kester, M. Dysregulation of sphingolipid metabolism in cancer. Cancer Biol. Ther. 2011, 11, 138–149. [Google Scholar] [CrossRef]
- Moon, B.S.; Park, M.T.; Park, J.H.; Kim, S.W.; Lee, K.C.; An, G.I.; Yang, S.D.; Chi, D.Y.; Cheon, G.J.; Lee, S.-J. Synthesis of novel phytosphingosine derivatives and their preliminary biological evaluation for enhancing radiation therapy. Bioorg. Med. Chem. Lett. 2007, 17, 6643–6646. [Google Scholar] [CrossRef] [PubMed]
- Park, M.T.; Kang, Y.H.; Park, I.C.; Kim, C.H.; Lee, Y.S.; Chung, H.Y.; Lee, S.-J. Combination treatment with arsenic trioxide and phytosphingosine enhances apoptotic cell death in arsenic trioxide-resistant cancer cells. Mol. Cancer Ther. 2007, 6, 82–92. [Google Scholar] [CrossRef] [Green Version]
- Kim, S. Method for Preparing Phytosphingosine Liposome Composition. US Patent 0104774, 10 May 2007. [Google Scholar]
- Başpınar, Y.; Isar, S.; Şahin, Y.; Akbaba, H.; Nalbantsoy, A.; Akbaba, G.E. Long-term Stability of Cationic Phytosphingosine Nanoemulsions as Delivery Systems for plasmid DNA. Celal Bayar Univ. J. Sci. 2022, 18, 107–118. [Google Scholar] [CrossRef]
- Andersen, C. In Vivo Estimation of Water Content in Cerebral White Matter of Brain Tumour Patients and Normal Individuals: Towards A Quantitative Brain Oedema Definition. Acta Neurochir. 1997, 139, 249–256. [Google Scholar] [CrossRef]
- Banay-Schwartz, M.; Palkovits, M.; Lajtha, A. Heterogeneous Distribution of Functionally Important Amino Acids in Brain Areas of Adult and Aging Humans. Neurochem. Res. 1993, 18, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Brady, S.T.; Siegel, G.J.; Albers, R.W.; Price, D.L. Basic Neurochemistry: Principles of Molecular, Cellular and Medical Neurobiology, 8th ed.; Academic Press: Boston, MA, USA, 2012; pp. 91–187. [Google Scholar]
- Chavko, M.; Nemoto, E.M.; Melick, J.A. Regional Lipid Composition in the Rat Brain. Mol. Chem. Neuropathol. 1993, 18, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Ellis, C.E.; Murphy, E.J.; Mitchell, D.C.; Golovko, M.Y.; Scaglia, F.; Barcelo-Coblijn, G.C.; Nussbaum, R.L. Mitochondrial Lipid Abnormality and Electron Transport Chain Impairment in Mice Lacking Synuclein. Mol. Cell. Biol. 2005, 25, 10190–10201. [Google Scholar] [CrossRef] [Green Version]
- Norton, W.T.; Abe, T.; Poduslo, S.E.; DeVries, G.H. The Lipid Composition of Isolated Brain Cells and Axons. J. Neurosci. Res. 1975, 1, 57–75. [Google Scholar] [CrossRef]
- Scandroglio, F.; Venkata, J.K.; Loberto, N.; Prioni, S.; Schuchman, E.H.; Chigorno, V.; Prinetti, A.; Sonnino, S. Lipid Content of Brain, Brain Membrane Lipid Domains, and Neurons from Acid Sphingomyelinase Deficient Mice: Lipid Content and ASM Deficient Mice. J. Neurochem. 2008, 107, 329–338. [Google Scholar] [CrossRef] [Green Version]
- Sultan, F. Brain Evolution: Analysis of Mammalian Brain Architecture. Nature 2002, 415, 133–134. [Google Scholar] [CrossRef]
- Bhatia, B.; Hsieh, M.; Kenney, A.M.; Nahlé, Z. Mitogenic Sonic hedgehog signaling drives E2F1-dependent lipogenesis in progenitor cells and medulloblastoma. Oncogene 2011, 30, 587–600. [Google Scholar] [CrossRef] [Green Version]
- Roser, M.; Josic, D.; Kontou, M.; Mosetter, K.; Maurer, P.; Reutter, W. Metabolism of galactose in the brain and liver of rats and its conversion into glutamate and other amino acids. J. Neural Transm. 2009, 116, 131–139. [Google Scholar] [CrossRef]
- von Bartheld, C.S.; Bahney, J.; Herculano-Houzel, S. The search for true numbers of neurons and glial cells in the human brain: A review of 150 years of cell counting. J. Comp. Neurol. 2016, 524, 3865–3895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel mutations target distinct subgroups of medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becker, S.A.; Palsson, B.O. Context-specific metabolic networks are consistent with experiments. PLoS Comput. Biol. 2008, 4, 1000082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tech, K.; Deshmukh, M.; Gershon, T.R. Adaptations of energy metabolism during cerebellar neurogenesis are co-opted in medulloblastoma. Cancer Lett. 2015, 356, 268–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venneti, S.; Thompson, C.B. Metabolic Reprogramming in Brain Tumors. Annu. Rev. Pathol. Mech. Dis. 2017, 12, 515–545. [Google Scholar] [CrossRef]
- Dranoff, G.; Elion, G.B.; Friedman, H.S.; Campbell, G.L.; Bigner, D.D. Influence of Glutamine on the Growth of Human Glioma and Medulloblastoma in Culture. Cancer Res. 1985, 45, 4077–4081. [Google Scholar]
- Dunkl, V.; Cleff, C.; Stoffels, G.; Judov, N.; Sarikaya-Seiwert, S.; Law, I.; Bøgeskov, L.; Nysom, K.; Andersen, S.B.; Steiger, H.-J.; et al. The usefulness of dynamic O-(2-18F-fluoroethyl)-L-tyrosine PET in the clinical evaluation of brain tumors in children and adolescents. J. Nucl. Med. 2015, 56, 88–92. [Google Scholar] [CrossRef] [Green Version]
- Xin, Y.; Yue, X.; Li, H.; Li, Z.; Cai, H.; Choudhary, A.K.; Zhang, S.; Chugani, D.C.; Langhans, S.A. pet imaging of medulloblastoma with an 18 f-labeled tryptophan analogue in a transgenic mouse model. Sci. Rep. 2020, 10, 3800. [Google Scholar] [CrossRef] [Green Version]
- Gururangan, S.; Hwang, E.; Herndon, J.E.; Fuchs, H.; George, T.; Coleman, R.E. [18F] fluorodeoxyglucose-positron emission tomography in patients with medulloblastoma. Neurosurgery 2004, 55, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Haggart, C.R.; Bartell, J.A.; Saucerman, J.J.; Papin, J.A. Whole-genome metabolic network reconstruction and constraint-based modeling. Methods Enzymol. 2011, 500, 411–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaufman, D.E.; Smith, R.L. Direction Choice for Accelerated Convergence in Hit-and-Run Sampling. Oper. Res. 1998, 46, 84–95. [Google Scholar] [CrossRef]








| MB | SHH | WNT | GR3 | GR4 | Healthy | Exp. Results for MB | |
|---|---|---|---|---|---|---|---|
| Glucose uptake rate (R593 + R594) | 0.852 | 0.852 | 0.852 | 0.852 | 0.852 | 0.080 | |
| Lactate production rate (R11 + R56) | 1.67 | 1.67 | 1.67 | 1.67 | 1.67 | 0.011 | Twice of glucose uptake rate [17] |
| Oxidative PPP rate/glucose Uptake (R17 + R61/R593 + R594) | 0.084 | 0.084 | 0.084 | 0.085 | 0.084 | 0.055 | Higher than healthy brain [14,18,19] |
| Non-Oxidative PPP rate (R21 + R65) | 0.024 | 0.024 | 0.024 | 0.024 | 0.024 | 0.001 | Higher than healthy brain [14,18,19] |
| Oxidative metabolism (TCA) flux (Citrate production) (R25 + R69) | 0.043 | 0.043 | 0.043 | 0.043 | 0.043 | 0.120 | Lower than healthy brain [11] |
| Acetyl-CoA flux (R28 + R72) | 0.043 | 0.043 | 0.043 | 0.043 | 0.043 | 0.003 | Higher than healthy brain [20] |
| Glutamate Production (R96) | 0.074 | 0.074 | 0.074 | 0.074 | 0.073 | 0.056 | Higher than healthy brain [12] |
| Glutamate production/Glutamine Production (E4/E3) | 1.43 | 1.43 | 1.43 | 1.43 | 1.43 | No Data | 1.18–1.71 [21] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ozbek, I.I.; Ulgen, K.O. Identification of Therapeutic Targets for Medulloblastoma by Tissue-Specific Genome-Scale Metabolic Model. Molecules 2023, 28, 779. https://doi.org/10.3390/molecules28020779
Ozbek II, Ulgen KO. Identification of Therapeutic Targets for Medulloblastoma by Tissue-Specific Genome-Scale Metabolic Model. Molecules. 2023; 28(2):779. https://doi.org/10.3390/molecules28020779
Chicago/Turabian StyleOzbek, Ilkay Irem, and Kutlu O. Ulgen. 2023. "Identification of Therapeutic Targets for Medulloblastoma by Tissue-Specific Genome-Scale Metabolic Model" Molecules 28, no. 2: 779. https://doi.org/10.3390/molecules28020779
APA StyleOzbek, I. I., & Ulgen, K. O. (2023). Identification of Therapeutic Targets for Medulloblastoma by Tissue-Specific Genome-Scale Metabolic Model. Molecules, 28(2), 779. https://doi.org/10.3390/molecules28020779