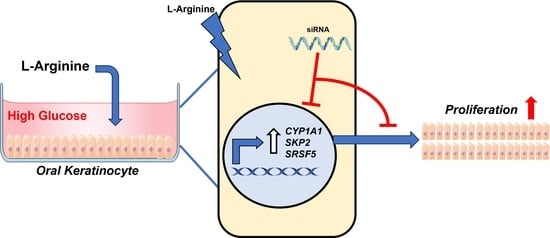
L-Arginine Enhances Oral Keratinocyte Proliferation under High-Glucose Conditions via Upregulation of CYP1A1, SKP2, and SRSF5
Abstract
:1. Introduction
2. Results
2.1. L-Arginine Enhances Oral Keratinocyte Proliferation
2.2. L-Arginine Leads to Significant Transcriptomic Changes in Oral Keratinocytes under High-Glucose Conditions
2.3. L-Arginine Induces Significant Protein Expression of CYP1A1, SRSF5, and SKP2 under High-Glucose Conditions
2.4. Knockdown of CYP1A1, SRSF5, and SKP2 Abolishes Enhanced Proliferation of Oral Keratinocytes by L-Arginine under High-Glucose Conditions
3. Materials and Methods
3.1. Cell Culture
3.2. RNA Sequencing and Bioinformatics Analysis
3.3. Indirect Immunofluorescent Cytochemistry
3.4. siRNA Knockdown of CYP1A1, SKP2, and SRSF5
3.5. Real-Time PCR
3.6. Statistical Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- National Diabetes Statistics Report; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2022.
- Diabetes; World Health Organization: Geneva, Switzerland, 2021.
- Retamal, I.; Hernández, R.; Velarde, V.; Oyarzún, A.; Martínez, C.; Julieta González, M.; Martínez, J.; Smith, P.C. Diabetes alters the involvement of myofibroblasts during periodontal wound healing. Oral Dis. 2020, 26, 1062–1071. [Google Scholar] [CrossRef]
- Devlin, H.; Garland, H.; Sloan, P. Healing of tooth extraction sockets in experimental diabetes mellitus. J. Oral. Maxillofac. Surg. 1996, 54, 1087–1091. [Google Scholar] [CrossRef]
- Brizeno, L.A.; Assreuy, A.M.; Alves, A.P.; Sousa, F.B.; de B Silva, P.G.; de Sousa, S.C.O.M.; Lascane, N.A.; Evangelista, J.S.; Mota, M.R. Delayed healing of oral mucosa in a diabetic rat model: Implication of TNF-α, IL-1β and FGF-2. Life Sci. 2016, 155, 36–47. [Google Scholar] [CrossRef] [PubMed]
- Genco, R.J.; Borgnakke, W.S. Diabetes as a potential risk for periodontitis: Association studies. Periodontology 2000 2020, 83, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Manfredi, M.; McCullough, M.J.; Vescovi, P.; Al-Kaarawi, Z.M.; Porter, S.R. Update on diabetes mellitus and related oral diseases. Oral Dis. 2004, 10, 187–200. [Google Scholar] [CrossRef] [PubMed]
- Saremi, A.; Nelson, R.G.; Tulloch-Reid, M.; Hanson, R.L.; Sievers, M.L.; Taylor, G.W.; Shlossman, M.; Bennett, P.H.; Genco, R.; Knowler, W.C. Periodontal disease and mortality in type 2 diabetes. Diabetes Care 2005, 28, 27–32. [Google Scholar] [CrossRef]
- Shi, J.; Han, C.; Chen, D.; Trivedi, H.M.; Bangash, H.I.; Chen, L. High Glucose Induces Late Differentiation and Death of Human Oral Keratinocytes. Curr. Issues Mol. Biol. 2022, 44, 4015–4027. [Google Scholar] [CrossRef]
- Guo, S.; Dipietro, L.A. Factors affecting wound healing. J. Dent. Res. 2010, 89, 219–229. [Google Scholar] [CrossRef]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound Healing: A Cellular Perspective. Physiol. Rev. 2019, 99, 665–706. [Google Scholar] [CrossRef]
- Barbul, A. Arginine: Biochemistry, physiology, and therapeutic implications. JPEN J. Parenter. Enteral Nutr. 1986, 10, 227–238. [Google Scholar] [CrossRef]
- Luiking, Y.C.; Deutz, N.E. Exogenous arginine in sepsis. Crit. Care Med. 2007, 35 (Suppl. 9), S557–S563. [Google Scholar] [CrossRef] [PubMed]
- Seifter, E.; Rettura, G.; Barbul, A.; Levenson, S.M. Arginine: An essential amino acid for injured rats. Surgery 1978, 84, 224–230. [Google Scholar]
- Stechmiller, J.K.; Childress, B.; Cowan, L. Arginine supplementation and wound healing. Nutr. Clin. Pract. 2005, 20, 52–61. [Google Scholar] [CrossRef]
- Witte, M.B.; Barbul, A. Arginine physiology and its implication for wound healing. Wound Repair. Regen. 2003, 11, 419–423. [Google Scholar] [CrossRef]
- De Jonge, W.J.; Kwikkers, K.L.; te Velde, A.A.; van Deventer, S.J.; Nolte, M.A.; Mebius, R.E.; Ruijter, J.M.; Lamers, M.C.; Lamers, W.H. Arginine deficiency affects early B cell maturation and lymphoid organ development in transgenic mice. J. Clin. Investig. 2002, 110, 1539–1548. [Google Scholar] [CrossRef] [PubMed]
- Gao, T.; Zhao, M.M.; Li, Y.J.; Zhang, L.; Li, J.L.; Yu, L.L.; Gao, F.; Zhou, G.H. Effects of in ovo feeding of L-arginine on the development of digestive organs, intestinal function and post-hatch performance of broiler embryos and hatchlings. J. Anim. Physiol. Anim. Nutr. 2018, 102, e166–e175. [Google Scholar] [CrossRef]
- Barbul, A.; Lazarou, S.A.; Efron, D.T.; Wasserkrug, H.L.; Efron, G. Arginine enhances wound healing and lymphocyte immune responses in humans. Surgery 1990, 108, 331–336. [Google Scholar] [PubMed]
- Witte, M.B.; Thornton, F.J.; Tantry, U.; Barbul, A. L-Arginine supplementation enhances diabetic wound healing: Involvement of the nitric oxide synthase and arginase pathways. Metabolism 2002, 51, 1269–1273. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Guo, Y.; Sheng, Q.; Shyr, Y. Advanced heat map and clustering analysis using heatmap3. Biomed. Res. Int. 2014, 2014, 986048. [Google Scholar] [CrossRef] [PubMed]
- Efron, D.T.; Kirk, S.J.; Regan, M.C.; Wasserkrug, H.L.; Barbul, A. Nitric oxide generation from L-arginine is required for optimal human peripheral blood lymphocyte DNA synthesis. Surgery 1991, 110, 327–334. [Google Scholar]
- Ochoa, J.B.; Strange, J.; Kearney, P.; Gellin, G.; Endean, E.; Fitzpatrick, E. Effects of L-arginine on the proliferation of T lymphocyte subpopulations. JPEN J. Parenter. Enteral Nutr. 2001, 25, 23–29. [Google Scholar] [CrossRef]
- Greene, J.M.; Feugang, J.M.; Pfeiffer, K.E.; Stokes, J.V.; Bowers, S.D.; Ryan, P.L. L-Arginine enhances cell proliferation and reduces apoptosis in human endometrial RL95-2 cells. Reprod. Biol. Endocrinol. 2013, 11, 15. [Google Scholar] [CrossRef]
- Fujiwara, T.; Kanazawa, S.; Ichibori, R.; Tanigawa, T.; Magome, T.; Shingaki, K.; Miyata, S.; Tohyama, M.; Hosokawa, K. L-arginine stimulates fibroblast proliferation through the GPRC6A-ERK1/2 and PI3K/Akt pathway. PLoS ONE 2014, 9, e92168. [Google Scholar] [CrossRef]
- Gallala, H.; Macheleidt, O.; Doering, T.; Schreiner, V.; Sandhoff, K. Nitric oxide regulates synthesis of gene products involved in keratinocyte differentiation and ceramide metabolism. Eur. J. Cell Biol. 2004, 83, 667–679. [Google Scholar] [CrossRef]
- Lenis, Y.Y.; Elmetwally, M.A.; Maldonado-Estrada, J.G.; Bazer, F.W. Physiological importance of polyamines. Zygote 2017, 25, 244–255. [Google Scholar] [CrossRef] [PubMed]
- Mendes, D.A.; Horinouchi, C.D.; Prudente Ada, S.; Soley Bda, S.; Assreuy, J.; Otuki, M.F.; Cabrini, D.A. In vivo participation of nitric oxide in hyperproliferative epidermal phenomena in mice. Eur. J. Pharmacol. 2012, 687, 1–8. [Google Scholar] [CrossRef]
- Hu, S.C.; Lan, C.E. High-glucose environment disturbs the physiologic functions of keratinocytes: Focusing on diabetic wound healing. J. Dermatol. Sci. 2016, 84, 121–127. [Google Scholar] [CrossRef]
- Lan, C.C.; Liu, I.H.; Fang, A.H.; Wen, C.H.; Wu, C.S. Hyperglycaemic conditions decrease cultured keratinocyte mobility: Implications for impaired wound healing in patients with diabetes. Br. J. Dermatol. 2008, 159, 1103–1115. [Google Scholar] [CrossRef] [PubMed]
- Spravchikov, N.; Sizyakov, G.; Gartsbein, M.; Accili, D.; Tennenbaum, T.; Wertheimer, E. Glucose effects on skin keratinocytes: Implications for diabetes skin complications. Diabetes 2001, 50, 1627–1635. [Google Scholar] [CrossRef] [PubMed]
- Terashi, H.; Izumi, K.; Deveci, M.; Rhodes, L.M.; Marcelo, C.L. High glucose inhibits human epidermal keratinocyte proliferation for cellular studies on diabetes mellitus. Int. Wound J. 2005, 2, 298–304. [Google Scholar] [CrossRef]
- Kalmes, M.; Hennen, J.; Clemens, J.; Blömeke, B. Impact of aryl hydrocarbon receptor (AhR) knockdown on cell cycle progression in human HaCaT keratinocytes. Biol. Chem. 2011, 392, 643–651. [Google Scholar] [CrossRef]
- Ding, L.; Li, R.; Sun, R.; Zhou, Y.; Zhou, Y.; Han, X.; Cui, Y.; Wang, W.; Lv, Q.; Bai, J. S-phase kinase-associated protein 2 promotes cell growth and motility in osteosarcoma cells. Cell Cycle 2017, 16, 1547–1555. [Google Scholar] [CrossRef]
- Hershko, D.D. Oncogenic properties and prognostic implications of the ubiquitin ligase SKP2 in cancer. Cancer 2008, 112, 1415–1424. [Google Scholar] [CrossRef]
- Long, J.C.; Caceres, J.F. The SR protein family of splicing factors: Master regulators of gene expression. Biochem. J. 2009, 417, 15–27. [Google Scholar] [CrossRef]
- Huang, C.S.; Shen, C.Y.; Wang, H.W.; Wu, P.E.; Cheng, C.W. Increased expression of SRp40 affecting CD44 splicing is associated with the clinical outcome of lymph node metastasis in human breast cancer. Clin. Chim. Acta 2007, 384, 69–74. [Google Scholar] [CrossRef]
- Kim, H.R.; Lee, G.O.; Choi, K.H.; Kim, D.K.; Ryu, J.S.; Hwang, K.E.; Na, K.J.; Choi, C.; Kuh, J.H.; Chung, M.J.; et al. SKP2: A novel marker for small-cell lung cancer and pleural metastatic cancer. Lung Cancer 2016, 99, 57–65. [Google Scholar] [CrossRef]
- Piekielko-Witkowska, A.; Wiszomirska, H.; Wojcicka, A.; Poplawski, P.; Boguslawska, J.; Tanski, Z.; Nauman, A. Disturbed expression of splicing factors in renal cancer affects alternative splicing of apoptosis regulators, oncogenes, and tumor suppressors. PLoS ONE 2010, 5, e13690. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Jia, R.; Bian, Z. SKP2 functions as a novel oncogenic splicing factor and is upregulated by oncogene SRSF3 in oral squamous cell carcinoma. Biochim. Biophys. Acta Mol. Cell Res. 2018, 1865, 1161–1172. [Google Scholar] [CrossRef]
- Jensen, M.A.; Wilkinson, J.E.; Krainer, A.R. Splicing factor SRSF6 promotes hyperplasia of sensitized skin. Nat. Struct. Mol. Biol. 2014, 21, 189–197. [Google Scholar] [CrossRef]
- Barbul, A.; Fishel, R.S.; Shimazu, S.; Wasserkrug, H.L.; Yoshimura, N.N.; Tao, R.C.; Efron, G. Intravenous hyperalimentation with high arginine levels improves wound healing and immune function. J. Surg. Res. 1985, 38, 328–334. [Google Scholar] [CrossRef]
- De Luis, D.A.; Izaola, O.; Cuellar, L.; Terroba, M.C.; Martin, T.; Aller, R. High dose of arginine enhanced enteral nutrition in postsurgical head and neck cancer patients. A randomized clinical trial. Eur. Rev. Med. Pharmacol. Sci. 2009, 13, 279–283. [Google Scholar] [PubMed]
- De Luis, D.A.; Izaola, O.; Cuellar, L.; Terroba, M.C.; Martin, T.; Ventosa, M. A randomized double-blind clinical trial with two different doses of arginine enhanced enteral nutrition in postsurgical cancer patients. Eur. Rev. Med. Pharmacol. Sci. 2010, 14, 941–945. [Google Scholar]
- Ben Lagha, A.; Yang, Y.; Trivedi, H.M.; Masters, J.G.; Grenier, D. A Dual Zinc plus Arginine formulation protects against tumor necrosis factor-alpha-induced barrier dysfunction and enhances cell proliferation and migration in an in vitro gingival keratinocyte model. Arch. Oral. Biol. 2021, 126, 105126. [Google Scholar] [CrossRef] [PubMed]
- Malone-Povolny, M.J.; Maloney, S.E.; Schoenfisch, M.H. Nitric Oxide Therapy for Diabetic Wound Healing. Adv. Healthc. Mater. 2019, 8, e1801210. [Google Scholar] [CrossRef]
- Schäffer, M.R.; Tantry, U.; Efron, P.A.; Ahrendt, G.M.; Thornton, F.J.; Barbul, A. Diabetes-impaired healing and reduced wound nitric oxide synthesis: A possible pathophysiologic correlation. Surgery 1997, 121, 513–519. [Google Scholar] [CrossRef]
- Shi, H.P.; Most, D.; Efron, D.T.; Witte, M.B.; Barbul, A. Supplemental L-arginine enhances wound healing in diabetic rats. Wound Repair. Regen. 2003, 11, 198–203. [Google Scholar] [CrossRef] [PubMed]
- Zandifar, A.; Seifabadi, S.; Zandifar, E.; Beheshti, S.S.; Aslani, A.; Javanmard, S.H. Comparison of the effect of topical versus systemic L-arginine on wound healing in acute incisional diabetic rat model. J. Res. Med. Sci. 2015, 20, 233–238. [Google Scholar]
- Arana, V.; Paz, Y.; González, A.; Méndez, V.; Méndez, J.D. Healing of diabetic foot ulcers in L-arginine-treated patients. Biomed. Pharmacother. 2004, 58, 588–597. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.G.; Hanft, J.R.; Driver, V.R.; Smith, A.P.; Lazaro-Martinez, J.L.; Reyzelman, A.M.; Furst, G.J.; Vayser, D.J.; Cervantes, H.L.; Snyder, R.J.; et al. Effect of oral nutritional supplementation on wound healing in diabetic foot ulcers: A prospective randomized controlled trial. Diabet. Med. 2014, 31, 1069–1077. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, J.; Leonardo, T.R.; Han, C.; Bangash, H.I.; Chen, D.; Trivedi, H.M.; Chen, L. L-Arginine Enhances Oral Keratinocyte Proliferation under High-Glucose Conditions via Upregulation of CYP1A1, SKP2, and SRSF5. Molecules 2023, 28, 7020. https://doi.org/10.3390/molecules28207020
Shi J, Leonardo TR, Han C, Bangash HI, Chen D, Trivedi HM, Chen L. L-Arginine Enhances Oral Keratinocyte Proliferation under High-Glucose Conditions via Upregulation of CYP1A1, SKP2, and SRSF5. Molecules. 2023; 28(20):7020. https://doi.org/10.3390/molecules28207020
Chicago/Turabian StyleShi, Junhe, Trevor R. Leonardo, Chen Han, Hiba I. Bangash, Dandan Chen, Harsh M. Trivedi, and Lin Chen. 2023. "L-Arginine Enhances Oral Keratinocyte Proliferation under High-Glucose Conditions via Upregulation of CYP1A1, SKP2, and SRSF5" Molecules 28, no. 20: 7020. https://doi.org/10.3390/molecules28207020
APA StyleShi, J., Leonardo, T. R., Han, C., Bangash, H. I., Chen, D., Trivedi, H. M., & Chen, L. (2023). L-Arginine Enhances Oral Keratinocyte Proliferation under High-Glucose Conditions via Upregulation of CYP1A1, SKP2, and SRSF5. Molecules, 28(20), 7020. https://doi.org/10.3390/molecules28207020