Integration of Deep Learning and Sequential Metabolism to Rapidly Screen Dipeptidyl Peptidase (DPP)-IV Inhibitors from Gardenia jasminoides Ellis
Abstract
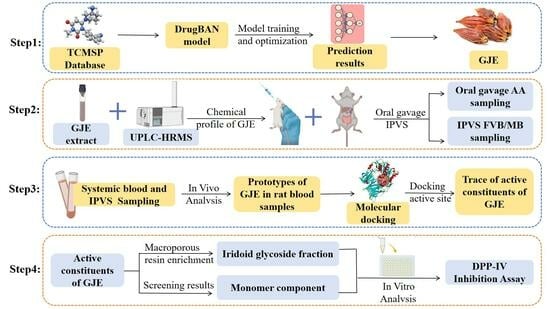
:1. Introduction
2. Results and Discussion
2.1. Validation of Deep-Learning Model
2.2. Deep-Learning Model Prediction and Filters
2.3. Identification of the Absorbed Components in Gardenia jasminoides Ellis
2.4. Molecular Docking Studies
2.5. In Vitro Activity Assay
3. Materials and Methods
3.1. Materials
3.2. Deep-Learning Model Predicts Compound Affinity for DPP-IV
3.2.1. Data Collection and Preparation
3.2.2. Deep-Learning Model
3.2.3. Model Optimization and Evaluation
3.2.4. Model Prediction
3.3. Preparation of Sample Solutions
3.4. Enrichment of the Iridoid Glycoside Extract of GJE with Macroporous Resin
3.5. Animals
3.6. Animal Experiments
3.6.1. In Vivo Metabolic Experiments
3.6.2. Intragastric Administration
3.7. UPLC-Q Exactive-Orbitrap HRMS Analysis
3.8. Molecular Docking
3.9. In Vitro DPP-IV Inhibition Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Putta, S.; Yarla, N.S.; Kilari, E.K.; Surekha, C.; Aliev, G.; Divakara, M.B.; Santosh, M.S.; Ramu, R.; Zameer, F.; Prasad, M.N.N.; et al. Therapeutic Potentials of Triterpenes in Diabetes and its Associated Complications. Curr. Top. Med. Chem. 2016, 16, 2532–2542. [Google Scholar] [CrossRef]
- Samandari, R.; Chizari, A.; Hassanpour, R.; Mousavi, Z.; Haghparast, A. Streptozotocin-induced diabetes affects the development morphine reward in rats. Neurosci. Lett. 2013, 543, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Vorsanger, M.H.; Subramanyam, P.; Weintraub, H.S.; Lamm, S.H.; Underberg, J.A.; Gianos, E.; Goldberg, I.J.; Schwartzbard, A.Z. Cardiovascular Effects of the New Weight Loss Agents. J. Am. Coll. Cardiol. 2016, 68, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Nauck, M. Incretin therapies: Highlighting common features and differences in the modes of action of glucagon-like peptide-1 receptor agonists and dipeptidyl peptidase-4 inhibitors. Diabetes Obes. Metab. 2016, 18, 203–216. [Google Scholar] [CrossRef] [PubMed]
- Kerr, B.D.; Flatt, P.R.; Gault, V.A. Effects of gamma-glutamyl linker on DPP-IV resistance, duration of action and biological efficacy of acylated glucagon-like peptide-1. Biochem. Pharmacol. 2010, 80, 396–401. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.D. Potential for combination of dipeptidyl peptidase-4 inhibitors and sodium-glucose co-transporter-2 inhibitors for the treatment of type 2 diabetes. Diabetes Obes. Metab. 2015, 17, 616–621. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Yadav, D.; Sharma, N.; Jin, J.O. Dipeptidyl Peptidase (DPP)-IV Inhibitors with Antioxidant Potential Isolated from Natural Sources: A Novel Approach for the Management of Diabetes. Pharmaceuticals 2021, 14, 586. [Google Scholar] [CrossRef]
- Li, C.J.; Liu, X.J.; Bai, L.; Yu, Q.; Zhang, Q.M.; Yu, P.; Yu, D.M. Efficacy and safety of vildagliptin, Saxagliptin or Sitagliptin as add-on therapy in Chinese patients with type 2 diabetes inadequately controlled with dual combination of traditional oral hypoglycemic agents. Diabetol. Metab. Syndr. 2014, 6, 69. [Google Scholar] [CrossRef]
- Howse, P.M.; Chibrikova, L.N.; Twells, L.K.; Barrett, B.J.; Gamble, J.M. Safety and Efficacy of Incretin-Based Therapies in Patients With Type 2 Diabetes Mellitus and CKD: A Systematic Review and Meta-analysis. Am. J. Kidney Dis. 2016, 68, 733–742. [Google Scholar] [CrossRef]
- Shu, Y.S.; He, D.; Li, W.; Wang, M.L.; Zhao, S.Y.; Liu, L.L.; Cao, Z.W.; Liu, R.; Huang, Y.J.; Li, H.; et al. Hepatoprotective Effect of Citrus aurantium L. Against APAP-induced Liver Injury by Regulating Liver Lipid Metabolism and Apoptosis. Int. J. Biol. Sci. 2020, 16, 752–765. [Google Scholar] [CrossRef]
- Li, L.S.; Zhang, J.L.; Qiao, Q.H.; Wu, L.H.; Chen, L.Y. Development, Reliability, and Validity of the “Knowledge-Attitude-Practice” Questionnaire of Foreigners on Traditional Chinese Medicine Treatment. Evid.-Based Complement. Altern. Med. 2020, 2020, 8527320. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.X.; Chen, C.; Fu, X. Hypoglycemic activity in vitro and vivo of a water-soluble polysaccharide from Astragalus membranaceus. Food Funct. 2022, 13, 11210–11222. [Google Scholar] [CrossRef] [PubMed]
- Li, J.S.; Ji, T.; Su, S.L.; Zhu, Y.; Chen, X.L.; Shang, E.X.; Guo, S.; Qian, D.W.; Duan, J.A. Mulberry leaves ameliorate diabetes via regulating metabolic profiling and AGEs/RAGE and p38 MAPK/NF-kappa B pathway. J. Ethnopharmacol. 2022, 283, 114713. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.M.; Zhang, Y.L.; Peng, Y.; Li, X.B. The water extract of Radix scutellariae, its total flavonoids and baicalin inhibited CYP7A1 expression, improved bile acid, and glycolipid metabolism in T2DM mice. J. Ethnopharmacol. 2022, 293, 115238. [Google Scholar] [CrossRef]
- Shi, J.N.; Hu, H.; Harnett, J.; Zheng, X.T.; Liang, Z.J.; Wang, Y.T.; Ung, C.O.L. An evaluation of randomized controlled trials on nutraceuticals containing traditional Chinese medicines for diabetes management: A systematic review. Chin. Med. 2019, 14, 54. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.H.; Zhang, J.; Li, C.Q.; Zou, L.W. Discovery of anthraquinones as DPP-IV inhibitors: Structure-activity relationships and inhibitory mechanism. Fitoterapia 2023, 168, 105549. [Google Scholar] [CrossRef]
- Rifaioglu, A.S.; Atas, H.; Martin, M.J.; Cetin-Atalay, R.; Atalay, V.; Dogan, T. Recent applications of deep learning and machine intelligence on in silico drug discovery: Methods, tools and databases. Brief. Bioinform. 2019, 20, 1878–1912. [Google Scholar] [CrossRef]
- Xue, H.; Li, J.; Xie, H.; Wang, Y. Review of Drug Repositioning Approaches and Resources. Int. J. Biol. Sci. 2018, 14, 1232–1244. [Google Scholar] [CrossRef]
- Cheuka, P.M.; Mayoka, G.; Mutai, P.; Chibale, K. The Role of Natural Products in Drug Discovery and Development against Neglected Tropical Diseases. Molecules 2017, 22, 58. [Google Scholar] [CrossRef]
- Peska, L.; Buza, K.; Koller, J. Drug-target interaction prediction: A Bayesian ranking approach. Comput. Methods Programs Biomed. 2017, 152, 15–21. [Google Scholar] [CrossRef]
- Ezzat, A.; Wu, M.; Li, X.L.; Kwoh, C.K. Computational prediction of drug-target interactions using chemogenomic approaches: An empirical survey. Brief. Bioinform. 2019, 20, 1337–1357. [Google Scholar] [CrossRef]
- Perez-Lopez, C.; Molina, A.; Lozoya, E.; Segarra, V.; Municoy, M.; Guallar, V. Combining machine-learning and molecular-modeling methods for drug-target affinity predictions. Wiley Interdiscip. Rev.-Comput. Mol. Sci. 2023, 13, e1653. [Google Scholar] [CrossRef]
- Bagherian, M.; Sabeti, E.; Wang, K.; Sartor, M.A.; Nikolovska-Coleska, Z.; Najarian, K. Machine learning approaches and databases for prediction of drug-target interaction: A survey paper. Brief. Bioinform. 2021, 22, 247–269. [Google Scholar] [CrossRef] [PubMed]
- Li, J.J.; Li, L.; Wang, Y.Y.; Zhao, Y.X.; Hu, P.; Xu, Z.; Liu, F.; Liang, Q.Q.; Tian, X.T.; Huang, C.G. Systematic investigation on the anti-rheumatoid arthritis material basis and mechanism of Juan Bi Tang. Part 1: Integrating metabolic profiles and network pharmacology. J. Pharm. Biomed. Anal. 2021, 202, 114133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.L.; He, S.R.; Wu, S.Q.; Li, Y.; Wang, H.Y.; Yan, C.Y.; Yang, H.; Li, P. Discovering a Multi-Component Combination against Vascular Dementia from Danshen-Honghua Herbal Pair by Spectrum-Effect Relationship Analysis. Pharmaceuticals 2022, 15, 1073. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.T.; Xu, Z.; Chen, M.C.; Hu, P.; Liu, F.; Sun, Z.L.; Liu, H.; Guo, X.Z.; Li, Z.X.; Huang, C.G. Simultaneous determination of eight bioactive compounds by LC-MS/MS and its application to the pharmacokinetics, liver first-pass effect, liver and brain distribution of orally administrated Gouteng-Baitouweng (GB) in rats. J. Chromatogr. B-Anal. Technol. Biomed. Life Sci. 2018, 1084, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Liao, M.L.; Shang, H.H.; Li, Y.Z.; Li, T.; Wang, M.; Zheng, Y.A.; Hou, W.B.; Liu, C.X. An integrated approach to uncover quality marker underlying the effects of Alisma orientale on lipid metabolism, using chemical analysis and network pharmacology. Phytomedicine 2018, 45, 93–104. [Google Scholar] [CrossRef]
- Luo, Z.Q.; Liu, Y.; Han, X.; Yang, W.N.; Wang, G.P.; Wang, J.; Jiang, X.Q.; Sen, M.L.; Li, X.Y.; Yu, G.H.; et al. Mechanism of Paeoniae Radix Alba in the Treatment of Non-alcoholic Fatty Liver Disease Based on Sequential Metabolites Identification Approach, Network Pharmacology, and Binding Affinity Measurement. Front. Nutr. 2021, 8, 677659. [Google Scholar] [CrossRef]
- Zhou, H.B.; Zhang, S.; Chen, L.H.; Liu, Y.M.; Shen, L.H.; Zhang, J.L. Effective Therapeutic Verification of Crocin I, Geniposide, and Gardenia (Gardenia jasminoides Ellis) on Type 2 Diabetes Mellitus In Vivo and In Vitro. Foods 2023, 12, 1668. [Google Scholar] [CrossRef]
- Ye, Z.Y.; Chen, X.; He, Y.L.; Jin, M.Z.; Ye, M. Antidiabetic effects of fermented milk contained with Gardenia jasminoides water extracts on streptozotocin-induced mice. J. Food Process. Preserv. 2021, 45, e14785. [Google Scholar] [CrossRef]
- Guan, L.L.; Feng, H.Y.; Gong, D.Z.; Zhao, X.; Cai, L.; Wu, Q.; Yuan, B.; Yang, M.; Zhao, J.; Zou, Y. Genipin ameliorates age-related insulin resistance through inhibiting hepatic oxidative stress and mitochondrial dysfunction. Exp. Gerontol. 2013, 48, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Miura, T.; Nishiyama, Y.; Ichimaru, M.; Moriyasu, M.; Kato, A. Hypoglycemic activity and structure-activity relationship of iridoidal glycosides. Biol. Pharm. Bull. 1996, 19, 160–161. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.Q.; Ma, X.Y.; Liu, Y.; Lu, L.N.; Yang, R.R.; Yu, G.H.; Sun, M.H.; Xin, S.K.; Tian, S.M.; Chen, X.J.; et al. An Approach to Characterizing the Complicated Sequential Metabolism of Salidroside in Rats. Molecules 2016, 21, 706. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Deng, H.; Liu, L.; Liu, S.; Ren, L.; Shi, C.; Chen, X.; Guan, L.; Liu, W.; Li, Z.; et al. Highly Accessible Computational Prediction and In Vivo/In Vitro Experimental Validation: Novel Synthetic Phenyl Ketone Derivatives as Promising Agents against NAFLD via Modulating Oxidoreductase Activity. Oxid. Med. Cell. Longev. 2023, 2023, 3782230. [Google Scholar] [CrossRef]
- Dong, M.; Vattelana, A.M.; Lam, P.C.H.; Orry, A.J.; Abagyan, R.; Christopoulos, A.; Sexton, P.M.; Haines, D.R.; Miller, L.J. Development of a Highly Selective Allosteric Antagonist Radioligand for the Type 1 Cholecystokinin Receptor and Elucidation of Its Molecular Basis of Binding. Mol. Pharmacol. 2015, 87, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, M.; Pan, F.; Li, J.; Dou, R.; Wang, X.; Wang, Y.; He, Y.; Wang, S.; Cai, S. analysis of novel dipeptidyl peptidase-IV inhibitory peptides released from antimicrobial protein 2 (MiAMP2) and the possible pathways involved in diabetes protection. Curr. Res. Food Sci. 2021, 4, 603–611. [Google Scholar] [CrossRef] [PubMed]
- Araki, M.; Kanegawa, N.; Iwata, H.; Sagae, Y.; Ito, K.; Masuda, K.; Okuno, Y. Hydrophobic interactions at subsite S1′ of human dipeptidyl peptidase IV contribute significantly to the inhibitory effect of tripeptides. Heliyon 2020, 6, e04227. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.; Skonberg, D.I.; Myracle, A.D. Anti-Hyperglycemic Effects of Green Crab Hydrolysates Derived by Commercially Available Enzymes. Foods 2020, 9, 258. [Google Scholar] [CrossRef]
- Bai, P.; Miljković, F.; John, B.; Lu, H. Interpretable bilinear attention network with domain adaptation improves drug-target prediction. Nat. Mach. Intell. 2023, 5, 126–136. [Google Scholar] [CrossRef]
- Luo, Z.Q.; Liu, Y.; Zhao, B.S.; Tang, M.M.; Dong, H.H.; Zhang, L.; Lv, B.R.; Wei, L. Ex vivo and in situ approaches used to study intestinal absorption. J. Pharmacol. Toxicol. Methods 2013, 68, 208–216. [Google Scholar] [CrossRef]
- Li, H.W.; Dong, L.; Liu, Y.; Wang, G.; Zhang, L.; Qiao, Y.J. Comparison of two approaches of intestinal absorption by puerarin. J. Pharmacol. Toxicol. Methods 2014, 70, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Patel, B.D.; Ghate, M.D. Recent approaches to medicinal chemistry and therapeutic potential of dipeptidyl peptidase-4 (DPP-4) inhibitors. Eur. J. Med. Chem. 2014, 74, 574–605. [Google Scholar] [CrossRef] [PubMed]
- Musoev, A.; Numonov, S.; You, Z.H.; Gao, H.W. Discovery of Novel DPP-IV Inhibitors as Potential Candidates for the Treatment of Type 2 Diabetes Mellitus Predicted by 3D QSAR Pharmacophore Models, Molecular Docking and De Novo Evolution. Molecules 2019, 24, 2870. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Soman, S.S. Design and synthesis of sulfonamide derivatives of pyrrolidine and piperidine as anti-diabetic agents. Eur. J. Med. Chem. 2015, 90, 342–350. [Google Scholar] [CrossRef]





| Metrics | Trained Model | Random Prediction |
|---|---|---|
| AUROC | 0.8810 | 0.5000 |
| AUPRC | 0.7319 | 0.2813 |
| F1 score | 0.9729 | - |
| Sensitivity | 0.9865 | - |
| Specificity | 0.9600 | - |
| Accuracy | 0.9832 | - |
| Threshold | 0.2706 | - |
| No. | Source of Compound | Compound’s Name | TCMSP ID | P27487 Score | P28843 Score | P14740 Score | Total Score |
|---|---|---|---|---|---|---|---|
| 1 | Ecliptae Herba | Ecliptasaponin D | MOL003383 | 0.9798 | 0.8247 | 0.8418 | 2.6463 |
| 2 | Xanthium sibiricum Patr. | Strumaroside | MOL002294 | 0.9796 | 0.8506 | 0.9227 | 2.7529 |
| 3 | Ephedra Herba | 2,6-Dimethyl-1,3,5,7-octatetraene, E,E- | MOL003935 | 0.9795 | 0.7253 | 0.8179 | 2.5227 |
| 4 | Magnolia Officinalis Rehd Et Wils. | 5-allyl-3-(4-allylphenoxy)pyrocatechol | MOL005972 | 0.9795 | 0.6784 | 0.8428 | 2.5007 |
| 5 | Myrrha | 3β-acetoxy-16β-hydroxydammar-24-ene | MOL001150 | 0.9793 | 0.9315 | 0.9425 | 2.8533 |
| 6 | Dictamni Cortex | Dictamnoside | MOL006254 | 0.9793 | 0.7955 | 0.8364 | 2.6112 |
| 7 | Radix Paeoniae Rubra | 1-O-β-d-glucopyranosyl-8-O-benzoylpaeonisuffrone | MOL006993 | 0.9793 | 0.6134 | 0.7151 | 2.3078 |
| 8 | Tribulifructus | Spirostanol | MOL008580 | 0.9792 | 0.7898 | 0.9191 | 2.6881 |
| 9 | Gardenia jasminoides Ellis | Genipin 1-gentiobioside | MOL009038 | 0.9792 | 0.7189 | 0.7703 | 2.4684 |
| 10 | Dysosmae Verspiellis Rhixoma Et Radix | (E)-4-[(1S)-2,6,6-trimethyl-1-cyclohex-2-enyl]but-3-en-2-one | MOL011707 | 0.9791 | 0.8813 | 0.886 | 2.7464 |
| 11 | A. Dahurica (Fisch.) Benth. Et Hook | Daturic acid | MOL011501 | 0.9788 | 0.844 | 0.8818 | 2.7046 |
| 12 | Polygonati Rhizoma | Sibiricoside B | MOL009762 | 0.9787 | 0.8104 | 0.8806 | 2.6697 |
| 13 | Gardenia jasminoides Ellis | Shanziside | MOL004560 | 0.9786 | 0.8053 | 0.8381 | 2.622 |
| 14 | Croci Stigma | Carthamin | MOL001413 | 0.9786 | 0.6905 | 0.8393 | 2.5084 |
| 15 | Ginkgo Semen | Amentoflavone | MOL012037 | 0.9786 | 0.9096 | 0.9011 | 2.7893 |
| 16 | Pulsatilliae Radix | 5,6,7-Trimethoxycoumarin | MOL011997 | 0.9786 | 0.9096 | 0.9011 | 2.7893 |
| 17 | Arum Ternatum Thunb. | (Z)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enoic acid | MOL010389 | 0.9784 | 0.8322 | 0.9062 | 2.7168 |
| 18 | Gardenia jasminoides Ellis | Gardenoside | MOL004554 | 0.9784 | 0.8015 | 0.7198 | 2.4997 |
| 19 | Hedysarum Multijugum Maxim. | Isoflavanone | MOL010398 | 0.9784 | 0.8322 | 0.9062 | 2.7168 |
| 20 | Imperatae Rhizoma | Bifendate | MOL010387 | 0.9784 | 0.8322 | 0.9062 | 2.7168 |
| 21 | Gardenia jasminoides Ellis | Geniposidic acid | MOL001668 | 0.9782 | 0.8552 | 0.8563 | 2.6897 |
| 22 | Hedysarum Multijugum Maxim. | AstragalosideⅢ_ | MOL010406 | 0.9782 | 0.8322 | 0.9062 | 2.7166 |
| 23 | Carthami Flos | Carthamin-precursor | MOL002779 | 0.9782 | 0.6842 | 0.838 | 2.5004 |
| 24 | Impatientis Semen | Hosenkosides C | MOL008609 | 0.9782 | 0.8973 | 0.9492 | 2.8247 |
| 25 | Isatidis Radix | 5-(methoxymethyl)-2-furoic acid | MOL011822 | 0.9782 | 0.8899 | 0.9447 | 2.8128 |
| 26 | Gardenia jasminoides Ellis | 1,8-dihydroxy-3-Methylol-9,10-anthraquinone | MOL010471 | 0.9776 | 0.6499 | 0.7134 | 2.3409 |
| 27 | Radix Cynanchi Paniculati | Tomentogenin | MOL005622 | 0.9774 | 0.9162 | 0.9443 | 2.8379 |
| 28 | Isatidis Radix | 3-[2′-(5′-hydroxymethyl)furyl]-1(2H)-isoquinolinone-7-O-β-d-glucoside | MOL011727 | 0.9774 | 0.8676 | 0.9538 | 2.7988 |
| 29 | Gardenia jasminoides Ellis | Scandoside | MOL003135 | 0.9780 | 0.8923 | 0.8477 | 2.718 |
| 30 | Eupatorium Fortunei Turcz | Taraxasteryl palmitate | MOL000605 | 0.9780 | 0.8676 | 0.9538 | 2.7994 |
| Peak No. | tR/min | Measured Mass | Error (ppm) | Molecular Formula | Prototypical Compounds | MB | FVB | AA |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.91 | [M − H]− 191.055 9 | 1.555 | C7H12O6 | Quinic acid | + | + | + |
| 2 | 0.98 | [M − H]− 173.045 3 | 1.512 | C7H10O5 | Shikimic acid | - | - | - |
| 3 | 2.65 | [M − H]− 391.124 5 | 1.134 | C16H24O11 | Shanzhiside isomers | + | + | + |
| 4 | 2.81 | [M − H]− 403.124 1 | 0.183 | C17H24O11 | Deacetylasperulosidic acid methyl ester | - | - | - |
| 5 | 2.84 | [M − H]− 389.108 7 | 0.883 | C16H22O11 | Scandoside | + | + | - |
| 6 | 2.98 | [M − H]− 373.113 7 | 0.639 | C16H22O10 | Gardoside * | + | + | - |
| 7 | 3.13 | [M − H]− 391.124 5 | 1.221 | C16H24O11 | Shanziside * | + | + | + |
| 8 | 3.32 | [M − H]− 403.124 1 | 1.101 | C17H24O11 | Gardenoside | + | + | + |
| 9 | 3.35 | [M − H]− 373.113 6 | 0.478 | C16H22O10 | Geniposidic acid * | + | + | + |
| 10 | 3.38 | [M − H]− 403.124 1 | −0.561 | C17H24O11 | Feretoside | + | + | + |
| 11 | 3.48 | [M − H]− 405.139 9 | 0.503 | C17H26O11 | Shanziside methyl ester * | + | + | + |
| 12 | 3.57 | [M − H]− 375.129 4 | 0.715 | C16H24O10 | Mussaenosidic acid | + | + | + |
| 13 | 3.73 | [M − H]− 345.155 2 | 0.775 | C16H26O8 | Jasminoside D | + | + | + |
| 14 | 3.96 | [M − H]− 327.144 7 | 0.832 | C16H24O7 | Zataroside B | + | + | + |
| 15 | 4.04 | [M − H]− 353.087 6 | 1.028 | C16H18O9 | 5/3-O-Caffeoyl-quinic acid | + | + | - |
| 16 | 4.2 | [M − H]− 549.181 5 | −0.883 | C23H34O15 | Genipin 1-gentiobioside | - | - | + |
| 17 | 4.87 | [M − H]− 387.129 4 | 0.693 | C17H24O10 | Geniposide * | + | + | + |
| 18 | 5.34 | [M − H]− 345.155 2 | 0.775 | C16H26O8 | Jasminoside B | + | + | - |
| 19 | 5.49 | [M − H]− 353.087 4 | 0.405 | C16H18O9 | Chlorogenic acid | + | + | - |
| 20 | 6.33 | [M − H]− 179.034 8 | 1.832 | C9H8O4 | Caffeic acid | + | + | - |
| 21 | 6.39 | [M − H]− 183.102 3 | 1.206 | C10H16O3 | Jasminodiol | + | + | + |
| 22 | 6.62 | [M − H]− 503.176 9 | 0.883 | C22H32O13 | 2-methyl-lerythritol-4-O-(6-O-transsinapoyl)-β-d-glucopyranoside | + | + | - |
| 23 | 7.01 | [M − H]− 359.134 7 | 1.372 | C16H24O9 | Ixoroside | + | + | + |
| 24 | 7.44 | [M − H]− 429.139 8 | 0.335 | C19H26O11 | 10-acetyl geniposide | + | + | - |
| 25 | 7.57 | [M − H]− 519.150 6 | 0.614 | C25H28O12 | 6′-O-trans-coumaroyl geniposidic acid | + | - | - |
| 26 | 8.24 | [M − H]− 551.176 8 | 0.643 | C26H32O13 | 6-O-trans-p-coumaroyl Gardenoside methyl ester | + | - | - |
| 27 | 9.89 | [M − H]− 491.213 3 | 0.954 | C22H36O12 | Jasminoside S/H/I | + | - | - |
| 28 | 10.16 | [M − H]− 579.172 1 | 1.174 | C27H32O14 | 6′-O-trans-sinapoyl gardoside | + | - | - |
| 29 | 10.43 | [M − H]− 565.192 4 | 0.574 | C27H34O13 | 11-(6-O-trans-sinapoylglucopyranosyl)gardendiol | + | - | - |
| 30 | 10.79 | [M − H]− 609.146 3 | 1.216 | C27H30O16 | Rutin | - | - | - |
| 31 | 11.6 | [M − H]− 465.101 8 | 0.938 | C21H20O12 | Isoquercitrin | - | - | - |
| 32 | 11.86 | [M − H]− 593.151 3 | 1.121 | C27H30O15 | Nicotiflorin | + | - | - |
| 33 | 12.12 | [M − H]− 755.240 8 | 1.306 | C34H44O19 | 6″-O-trans-sinapoylgenipin gentiobioside | + | - | - |
| 34 | 12.62 | [M − H]− 725.230 2 | 1.201 | C33H42O18 | 6″-O-trans-feruloyl genipin gentiobioside | + | - | - |
| 35 | 12.69 | [M − H]− 695.219 2 | 0.742 | C32H40O17 | 6″-O-trans-p-coumaroylge nipin gentiobioside | + | - | - |
| 36 | 13.08 | [M − H]− 551.213 3 | 0.742 | C27H36O12 | 6′-O-trans-sinapoyl Jasminoside L | + | - | - |
| 37 | 13.34 | [M − H]− 975.371 0 | 0.044 | C44H64O24 | trans-crocin Ⅰ/cis-crocin Ⅰ | - | - | - |
| 38 | 14.07 | [M − H]− 593.187 7 | 1.095 | C28H34O14 | 6′-O-sinapoylgeniposide | + | - | - |
| 39 | 14.64 | [M − H]− 515.119 1 | 0.251 | C25H24O12 | 3,5-Dicaffeoylquinic acid | + | + | - |
| 40 | 15.51 | [M − H]− 533.166 3 | 0.786 | C26H30O12 | 6′-O-p-coumaroylgeniposide | + | + | - |
| 41 | 15.67 | [M − H]− 659.162 1 | 1.366 | C31H32O16 | 3,4-dicaffeovl-5-(3-hydroxy-3-methyl glutaroyl) quinic acid | + | + | - |
| 42 | 16.44 | [M − H]− 559.145 5 | 0.616 | C27H28O13 | 3-caffeoyl-4-sinapoylquinate | + | + | - |
| 43 | 17.45 | [M − H]− 535.218 3 | 0.716 | C27H36O11 | 6′-O-trans-sinapoyl jasminoside A | - | - | - |
| 44 | 18.57 | [M − H]− 533.202 5 | 0.419 | C27H34O11 | 6′-O-trans-sinapoyl jasminoside C | + | - | - |
| 45 | 19.58 | [M − H]− 345.061 4 | 0.95 | C17H14O8 | 5,7,3′,4′-tetrahydroxy-6,8-dimethoxy flavone | + | - | - |
| 46 | 21.75 | [M − H]− 813.319 2 | 1.286 | C38H54O19 | Crocin II | - | - | - |
| Num | Name | Libdock Score |
|---|---|---|
| 1 | Genipin 1-gentiobioside | 142.425 |
| 2 | Shanzhiside | 142.425 |
| 3 | Sitagliptin | 136.846 |
| 4 | Gardenoside | 132.894 |
| 5 | Geniposidic acid | 127.404 |
| 6 | Shanzhiside methyl ester | 107.752 |
| 7 | Scandoside | 107.136 |
| UniProt ID | Protein | Organism |
|---|---|---|
| P27487 | Dipeptidyl peptidase 4 | Homo sapiens (human) |
| P28843 | Dipeptidyl peptidase 4 | Mus musculus (mouse) |
| P14740 | Dipeptidyl peptidase 4 | Rattus norvegicus (rat) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, H.; Yu, S.; Li, X.; Wang, X.; Qi, D.; Pan, F.; Chai, X.; Wang, Q.; Pan, Y.; Zhang, L.; et al. Integration of Deep Learning and Sequential Metabolism to Rapidly Screen Dipeptidyl Peptidase (DPP)-IV Inhibitors from Gardenia jasminoides Ellis. Molecules 2023, 28, 7381. https://doi.org/10.3390/molecules28217381
Liu H, Yu S, Li X, Wang X, Qi D, Pan F, Chai X, Wang Q, Pan Y, Zhang L, et al. Integration of Deep Learning and Sequential Metabolism to Rapidly Screen Dipeptidyl Peptidase (DPP)-IV Inhibitors from Gardenia jasminoides Ellis. Molecules. 2023; 28(21):7381. https://doi.org/10.3390/molecules28217381
Chicago/Turabian StyleLiu, Huining, Shuang Yu, Xueyan Li, Xinyu Wang, Dongying Qi, Fulu Pan, Xiaoyu Chai, Qianqian Wang, Yanli Pan, Lei Zhang, and et al. 2023. "Integration of Deep Learning and Sequential Metabolism to Rapidly Screen Dipeptidyl Peptidase (DPP)-IV Inhibitors from Gardenia jasminoides Ellis" Molecules 28, no. 21: 7381. https://doi.org/10.3390/molecules28217381
APA StyleLiu, H., Yu, S., Li, X., Wang, X., Qi, D., Pan, F., Chai, X., Wang, Q., Pan, Y., Zhang, L., & Liu, Y. (2023). Integration of Deep Learning and Sequential Metabolism to Rapidly Screen Dipeptidyl Peptidase (DPP)-IV Inhibitors from Gardenia jasminoides Ellis. Molecules, 28(21), 7381. https://doi.org/10.3390/molecules28217381