Metal and Ligand Effect on the Structural Diversity of Divalent Coordination Polymers with Mixed Ligands: Evaluation for Photodegradation
Abstract
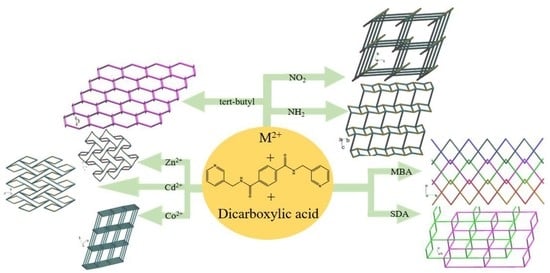
:1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Crystal Structure of 1
2.3. Crystal Structure of 2
2.4. Crystal Structure of 3
2.5. Crystal Structure of 4
2.6. Crystal Structures of 5
2.7. Crystal Structures of 6
2.8. Crystal Structures of 7
2.9. Crystal Structures of 8
2.10. Ligand Conformations and Coordination Modes
2.11. Structural Comparisons
2.12. Photodegradation
3. Materials and Methods
3.1. General Procedures
3.2. Materials
3.3. Preparations
3.3.1. [Co(L)(5-ter-IPA)(H2O)2]n, 1
3.3.2. {[Co(L)(5-NO2-IPA)]⋅2H2O}n, 2
3.3.3. {[Co(L)0.5(5-NH2-IPA)]⋅MeOH}n, 3
3.3.4. {[Co(L)(MBA)]⋅2H2O} n, 4
3.3.5. {[Co(L)(SDA)]⋅H2O}n , 5
3.3.6. {[Co2(L)2(1,4-NDC)2(H2O)2]⋅5H2O}n, 6
3.3.7. {[Cd(L)(1,4-NDC)(H2O)]⋅2H2O}n, 7
3.3.8. {[Zn2(L)2(1,4-NDC)2]⋅2H2O}n, 8
3.4. Powder X-ray Analysis and IR Spectra
3.5. Procedures for Photodegradation
3.6. X-ray Crystallography
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Lustig, W.P.; Mukherjee, S.; Rudd, N.D.; Desai, A.V.; Li, J.; Ghosh, S.K. Metal-organic frameworks: Functional luminescent and photonic materials for sensing applications. Chem. Soc. Rev. 2017, 46, 3242–3285. [Google Scholar] [CrossRef]
- Li, B.; Wen, H.-M.; Cui, Y.; Zhou, W.; Qian, G.; Chen, B. Emerging Multifunctional Metal-Organic Framework Materials. Adv. Mater. 2016, 28, 8819–8860. [Google Scholar] [CrossRef] [PubMed]
- Wales, D.J.; Grand, J.; Ting, V.P.; Burke, R.D.; Edler, K.J.; Bowen, C.R.; Mintova, S.; Burrows, A.D. Gas sensing using porous materials for automotive applications. Chem. Soc. Rev. 2015, 44, 4290–4321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kreno, L.E.; Leong, K.; Farha, O.K.; Allendorf, M.; Van Duyne, R.P.; Hupp, J.T. Metal–organic framework materials as chemical sensors. Chem. Rev. 2012, 112, 1105–1125. [Google Scholar] [CrossRef] [PubMed]
- Govindaraj, M.; Huang, W.-C.; Lee, C.-Y.; Lakshmanan, V.; Liu, Y.-H.; So, P.B.; Lin, C.-H.; Chen, J.-D. Structural Diversity of Mercury(II) Halide Complexes Containing Bis-pyridyl-bis-amide with Bulky and Angular Backbones: Ligand Effect and Metal Sensing. Int. J. Mol. Sci. 2022, 23, 7861. [Google Scholar] [CrossRef] [PubMed]
- Lakshmanan, V.; Lai, Y.-T.; Yang, X.-K.; Govindaraj, M.; Lin, C.-H.; Chen, J.-D. Eight-Fold Interpenetrating Diamondoid Coordination Polymers for Sensing Volatile Organic Compounds and Metal Ions. Polymers 2021, 13, 3018. [Google Scholar] [CrossRef]
- Chhetri, P.M.; Wu, M.-H.; Hsieh, C.-T.; Yang, X.-K.; Wu, C.-M.; Yang, E.-C.; Wang, C.-C.; Chen, J.-D. A Pair of CoII Supramolecular Isomers Based on Flexible Bis-Pyridyl-Bis-Amide and Angular Dicarboxylate Ligands. Molecules 2020, 25, 201. [Google Scholar] [CrossRef] [Green Version]
- Thapa, K.B.; Chen, J.-D. Crystal engineering of coordination polymers containing flexible bis-pyridyl-bis-amide ligands. CrystEngComm 2015, 17, 4611–4626. [Google Scholar] [CrossRef]
- Tang, S.; Gao, Y.; Han, S.; Chi, J.; Zhang, Z.; Liu, G. A kind of complex-based electrocatalytic sensor for monitoring the reduction of Cr(Ⅵ) by organic/inorganic reductants. Polyhedron 2022, 223, 115944. [Google Scholar] [CrossRef]
- Liu, G.; Han, S.; Gao, Y.; Xu, N.; Wang, X.; Chen, B. Multifunctional fluorescence responses of phenylamide-bridged d10 coordination polymers structurally regulated by dicarboxylates and metal ions. CrystEngComm 2020, 22, 7952–7961. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Li, J.-Y.; Li, J.-Q.; Xu, W.-Y.; Luo, M.-B.; Zhua, Y.; Luo, F. Exceptional temperature-dependent coordination sites from acylamide groups. Dalton Trans. 2014, 43, 5260–5264. [Google Scholar] [CrossRef] [PubMed]
- Blatov, V.A.; Shevchenko, A.P.; Proserpio, D.M. Applied Topological Analysis of Crystal Structures with the Program Package ToposPro. Cryst. Growth Des. 2014, 14, 3576–3586. [Google Scholar] [CrossRef]
- Bonneau, C.; O’Keeffe, M.; Proserpio, D.M.; Blatov, V.A.; Batten, S.R.; Bourne, S.A.; Lah, M.S.; Eon, J.G.; Hyde, S.T.; Wiggin, S.B.; et al. Deconstruction of Crystalline Networks into Underlying Nets: Relevance for Terminology Guidelines and Crystallographic Databases. Cryst. Growth Des. 2018, 18, 3411–3418. [Google Scholar] [CrossRef]
- Wen, T.; Zhang, D.-X.; Zhang, J. Two-Dimensional Copper(I) Coordination Polymer Materials as Photocatalysts for the Degradation of Organic Dyes. Inorg. Chem. 2013, 52, 12–14. [Google Scholar] [CrossRef]
- Wang, X.-L.; Luan, J.; Sui, F.-F.; Lin, H.-Y.; Liu, G.-C.; Xu, C. Structural Diversities and Fluorescent and Photocatalytic Properties of a Series of CuII Coordination Polymers Constructed from Flexible Bis-pyridyl-bis-amide Ligands with Different Spacer Lengths and Different Aromatic Carboxylates. Cryst. Growth Des. 2013, 13, 3561–3576. [Google Scholar] [CrossRef]
- Wu, Y.-P.; Wu, X.-Q.; Wang, J.-F.; Zhao, J.; Dong, W.-W.; Li, D.-S.; Zhang, Q.-C. Assembly of Two Novel Cd3/(Cd3 + Cd5)-Cluster-Based Metal−Organic Frameworks: Structures, Luminescence, and Photocatalytic Degradation of Organic Dyes. Cryst. Growth Des. 2016, 16, 2309–2316. [Google Scholar] [CrossRef]
- Cheng, H.-J.; Tang, X.-Y.; Yuan, R.-X.; Lang, J.-P. Structural diversity of Zn(II) coordination polymers based on bis-imidazolyl ligands and 5-R-1,3-benzenedicarboxylate and their photocatalytic properties. CrystEngComm 2016, 18, 4851–4862. [Google Scholar] [CrossRef]
- Li, D.-X.; Ren, Z.-G.; Young, D.J.; Lang, J.-P. Synthesis of Two Coordination Polymer Photocatalystsand Significant Enhancement of Their CatalyticPhotodegradation Activity by Doping with Co2+ Ions. Eur. J. Inorg. Chem. 2015, 11, 1981–1988. [Google Scholar] [CrossRef]
- Wang, X.-L.; Sha, X.-T.; Liu, G.-C.; Chen, N.-L.; Tian, Y. Polycarboxylate-directed various CoIJII) complexes based on a “V”-like bis-pyridyl-bis-amide derivative: Construction, electrochemical and photocatalytic properties. CrystEngComm 2015, 17, 7290–7299. [Google Scholar] [CrossRef]
- Chang, M.-N.; Yang, X.-K.; Chhetri, P.M.; Chen, J.-D. Metal and Ligand Effects on the Construction of Divalent Coordination Polymers Based on bis-pyridyl-bis-amide and Polycarboxylate Ligands. Polymers 2017, 9, 691. [Google Scholar] [CrossRef] [Green Version]
- Thapa, K.B.; Wu, M.-H.; Yang, X.-K.; Chen, T.-R.; Chen, J.-D. Co(II) coordination polymers exhibiting reversible structural transformation and color change: A comparative analysis with Ni(II) analogue. Polyhedron 2018, 152, 225–232. [Google Scholar] [CrossRef]
- Sumby, C.J.; Hanton, L.R. Syntheses and studies of flexible amide ligands: A toolkit for studying metallo-supramolecular assemblies for anion binding. Tetrahedron 2009, 65, 4681–4691. [Google Scholar] [CrossRef]
- Bruker AXS. APEX2, V2008.6, SADABS V2008/1, SAINT V7.60A, SHELXTL V6.14; Bruker AXS Inc.: Madison, WI, USA, 2008. [Google Scholar]
- Sheldrick, G.M. A short history of SHELX. Acta Crystallogr. 2008, A64, 112–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]










| Conformation | Coordination Mode | |
|---|---|---|
| 1 |  trans anti–anti  trans anti–anti |  μ2-κO:κO′ |
| 2 |  trans anti–anti  trans syn–syn |  μ3-κ2O,O′:κO′′:κO′′′ |
| 3 |  trans anti–anti |  μ4-κ2O,O′:κO′′:κO′′′:κN |
| 4 |  cis syn–syn |  μ2-κ2O,O′: κ2O′′,O′′′ |
| 5 |  trans anti–anti |  μ4-κO:κO′:κO′′:κO′′′ |
| 6 |  trans syn–syn  trans anti–anti |  μ2-κ2O,O′:κO′′ |
| 7 |  trans syn–syn |  μ2-κ2O,O′: κ2O′′,O′′′ |
| 8 |  trans syn–anti  cis anti–anti |  μ2-κO:κO′ |
| Compound | 1 | 2 | 3 |
|---|---|---|---|
| Formula | C32H34N4O8Co | C28H25N5O10Co | C19H18CoN3O6 |
| Formula weight | 661.56 | 650.46 | 443.29 |
| Crystal system | Triclinic | Triclinic | Triclinic |
| Space group | Pī | Pī | Pī |
| a, Å | 10.3115(3) | 10.0958(2) | 8.3283(3) |
| b, Å | 11.9684(4) | 11.3920(3) | 10.0484(3) |
| c, Å | 13.2748(4) | 12.6159(3) | 11.3130(4) |
| α, ° | 74.6853(9) | 73.5612(11) | 81.041(2) |
| β, ° | 73.1745(9) | 84.2220(10) | 87.225(25) |
| γ, ° | 73.1501(9) | 87.1266(11) | 82.0898(19) |
| V, Å3 | 1471.49(8) | 1384.23(6) | 925.94(5) |
| Z | 2 | 2 | 2 |
| Dcalc, Mg/m3 | 1.493 | 1.561 | 1.590 |
| F (000) | 690 | 670 | 456 |
| µ (Mo Kα), mm−1 | 0.644 | 0.689 | 0.971 |
| Range (2θ) for data collection, deg | 3.270 ≤ 2θ ≤ 56.854 | 3.380 ≤ 2θ ≤ 56.662 | 3.646 ≤ 2θ ≤ 56.688 |
| Independent reflections | 7357 [R(int) = 0.0247] | 6880 [R(int) = 0.0226] | 4607 [R(int) = 0.0329] |
| Data/restraints/parameters | 7357/0/406 | 6880/0/397 | 4607/0/262 |
| Quality-of-fit indicator c | 1.035 | 1.065 | 1.005 |
| Final R indices [I > 2σ(I)] a,b | R1 = 0.0303 wR2 = 0.0732 | R1 = 0.0536 wR2 = 0.1556 | R1 = 0.0378 wR2 = 0.0824 |
| R indices (all data) | R1 = 0.0369, wR2 = 0.0762 | R1 = 0.0656, wR2 = 0.1659 | R1 = 0.0548, wR2 = 0.0895 |
| Compound | 4 | 5 | 6 |
| Formula | C35H32CoN4O8 | C24H19CoN2O8S | C64H62Co2N8O19 |
| Formula weight | 695.57 | 554.40 | 1365.07 |
| Crystal system | Orthorhombic | Triclinic | Monoclinic |
| Space group | Ibca | Pī | P21/c |
| a, Å | 13.6153(3) | 9.1565(4) | 16.5329(3) |
| b, Å | 19.3800(5) | 10.9657(5) | 17.1867(3) |
| c, Å | 25.1236(6) | 13.2184(6) | 21.3467(4) |
| α, ° | 90 | 97.8126(15) | 90 |
| β, ° | 90 | 109.4504(17) | 103.3525(9) |
| γ, ° | 90 | 104.9062(15). | 90 |
| V, Å3 | 6629.2(3) | 1173.06(9) | 5901.61(19) |
| Z | 8 | 2 | 4 |
| Dcalc, Mg/m3 | 1.394 | 1.570 | 1.536 |
| F (000) | 2888 | 568 | 2832 |
| µ (Mo Kα), mm−1 | 0.576 | 0.874 | 0.649 |
| Range (2θ) for data collection, deg | 3.242 ≤ 2θ ≤ 52.000 | 3.962 ≤ 2θ ≤ 56.714 | 3.076 ≤ 2θ ≤ 56.588 |
| Independent reflections | 3267 [R(int) = 0.0767] | 5836 [R(int) = 0.0234] | 14613 [R(int) = 0.0466] |
| Data/restraints/parameters | 3267/1/223 | 5836/0/334 | 14613/0/841 |
| Quality-of-fit indicator c | 1.014 | 1.067 | 1.060 |
| Final R indices [I > 2σ(I)] a,b | R1 = 0.0492, wR2 = 0.1109 | R1 = 0.0375, wR2 = 0.1079 | R1 = 0.0526, wR2 = 0.1264 |
| R indices (all data) | R1 = 0.1309, wR2 = 0.1409 | R1 = 0.0493, wR2 = 0.1155 | R1 = 0.0892, wR2 = 0.1448 |
| Compound | 7 | 8 | |
| Formula | C32H30CdN4O9 | C64 H52Zn2N8O14 | |
| Formula weight | 727.00 | 1287.87 | |
| Crystal system | Monoclinic | Triclinic | |
| Space group | P21/c | Pī | |
| a, Å | 16.9732(3) | a = 12.8606(12) | |
| b, Å | 9.4163(2) | b = 13.1507(14) | |
| c, Å | 20.7184(4) | c = 18.7726(18) | |
| α, ° | 90 | 90.177(6) | |
| β, ° | 114.0812(10) | 101.414(6) | |
| γ, ° | 90 | 113.475(5) | |
| V, Å3 | 3023.12(10) | 2842.7(5) | |
| Z | 4 | 2 | |
| Dcalc, Mg/m3 | 1.597 | 1.505 | |
| F(000) | 1480 | 1328 | |
| µ(Mo Kα), mm−1 | 0.786 | 0.922 | |
| Range (2θ) for data collection, deg | 2.628 ≤ 2θ ≤ 56.620 | 2.222 ≤ 2θ ≤ 56.840 | |
| Independent reflections | 7521 [R(int) = 0.0571] | 14194 [R(int) = 0.0806] | |
| Data/restraints/parameters | 7521/0/419 | 14194/0/793 | |
| Quality-of-fit indicator c | 1.009 | 1.004 | |
| Final R indices [I > 2σ(I)] a,b | R1 = 0.0373, wR2 = 0.0670 | R1 = 0.0567, wR2 = 0.1012 | |
| R indices (all data) | R1 = 0.0672, wR2 = 0.0763 | R1 = 0.1485, wR2 = 0.1265 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Govindaraj, M.; Zhong, S.-Y.; Lin, C.-H.; Chen, J.-D. Metal and Ligand Effect on the Structural Diversity of Divalent Coordination Polymers with Mixed Ligands: Evaluation for Photodegradation. Molecules 2023, 28, 2226. https://doi.org/10.3390/molecules28052226
Govindaraj M, Zhong S-Y, Lin C-H, Chen J-D. Metal and Ligand Effect on the Structural Diversity of Divalent Coordination Polymers with Mixed Ligands: Evaluation for Photodegradation. Molecules. 2023; 28(5):2226. https://doi.org/10.3390/molecules28052226
Chicago/Turabian StyleGovindaraj, Manivannan, Shih-Ying Zhong, Chia-Her Lin, and Jhy-Der Chen. 2023. "Metal and Ligand Effect on the Structural Diversity of Divalent Coordination Polymers with Mixed Ligands: Evaluation for Photodegradation" Molecules 28, no. 5: 2226. https://doi.org/10.3390/molecules28052226
APA StyleGovindaraj, M., Zhong, S. -Y., Lin, C. -H., & Chen, J. -D. (2023). Metal and Ligand Effect on the Structural Diversity of Divalent Coordination Polymers with Mixed Ligands: Evaluation for Photodegradation. Molecules, 28(5), 2226. https://doi.org/10.3390/molecules28052226