Multiepitope Subunit Peptide-Based Nanovaccine against Porcine Circovirus Type 2 (PCV2) Elicited High Antibody Titers in Vaccinated Mice
Abstract
:1. Introduction
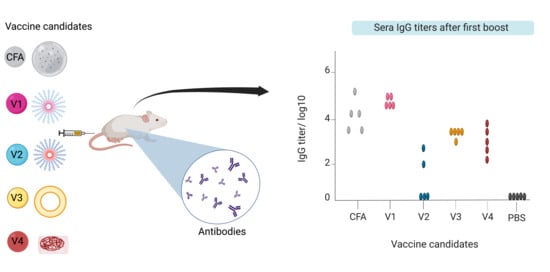
2. Results
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Equipment
4.3. Synthesis of Peptides, Conjugates and Formulations
4.4. Synthesis of Polymer Conjugates 5–8
4.5. Preparation of Vaccine Candidate V0
4.6. Preparation of Vaccine Candidate V1
4.7. Preparation of Vaccine Candidate V2
4.8. Preparation of Vaccine Candidate V3
4.9. Preparation of Vaccine Candidate V4
4.10. Dynamic Light Scattering
4.11. Transmission Electron Microscopy
4.12. Immunization
4.13. Antibody Titer Detection
4.14. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Segalés, J.; Domingo, M. Postweaning mulstisystemic wasting syndrome (PMWS) in pigs. A review. Vet. Q. 2002, 24, 109–124. [Google Scholar] [CrossRef]
- Franzo, G.; Segalés, J. Porcine circovirus 2 (PCV-2) genotype update and proposal of a new genotyping methodology. PLoS ONE 2018, 13, e0208585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, D.; Lefebvre, D.J.; Ooms, K.; Huang, L.; Delputte, P.L.; van Doorsselaere, J.; Nauwynck, H. Single amino acid mutations in the capsid switch the neutralization phenotype of porcine circovirus 2. J. Gen. Virol. 2012, 93, 1548–1555. [Google Scholar] [CrossRef]
- Gava, D.; Serrão, V.H.B.; Fernandes, L.T.; Cantão, M.E.; Ciacci-Zanella, J.R.; Morés, N.; Schaefer, R. Structure analysis of capsid protein of Porcine circovirus type 2 from pigs with systemic disease. Braz. J. Microbiol. 2018, 49, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Opriessnig, T.; Xiao, C.-T.; Gerber, P.F.; Halbur, P. Emergence of a novel mutant PCV2b variant associated with clinical PCVAD in two vaccinated pig farms in the US concurrently infected with PPV2. Veter. Microbiol. 2013, 163, 177–183. [Google Scholar] [CrossRef] [PubMed]
- Nawagitgul, P.; Harms, P.A.; Morozov, I.; Thacker, B.J.; Sorden, S.D.; Lekcharoensuk, C.; Paul, P. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin. Diagn. Lab. Immunol. 2002, 9, 33–40. [Google Scholar] [CrossRef] [Green Version]
- Blanchard, P.; Mahé, D.; Cariolet, R.; Keranflec’h, A.; Baudouard, M.A.; Cordioli, P.; Albina, E.; Jestin, A. Protection of swine against post-weaning multisystemic wasting syndrome (PMWS) by porcine circovirus type 2 (PCV2) proteins. Vaccine 2003, 21, 4565–4575. [Google Scholar] [CrossRef]
- Bandrick, M.; Gutiérrez, A.H.; Desai, P.; Rincon, G.; Martin, W.D.; Terry, F.E.; De Groot, A.S.; Foss, D.L. T cell epitope content comparison (EpiCC) analysis demonstrates a bivalent PCV2 vaccine has greater T cell epitope overlap with field strains than monovalent PCV2 vaccines. Vet. Immunol. Immunopathol. 2020, 223, 110034. [Google Scholar] [CrossRef]
- Karuppannan, A.K.; Opriessnig, T.J. Porcine circovirus type 2 (PCV2) vaccines in the context of current molecular epidemiology. Viruses 2017, 9, 99. [Google Scholar] [CrossRef] [Green Version]
- Bandrick, M.; Balasch, M.; Heinz, A.; Taylor, L.; King, V.; Toepfer, J.; Foss, D. A bivalent porcine circovirus type 2 (PCV2), PCV2a-PCV2b, vaccine offers biologically superior protection compared to monovalent PCV2 vaccines. Vet. Res. 2022, 53, 12. [Google Scholar] [CrossRef]
- Saha, D.; Huang, L.; Bussalleu, E.; Lefebvre, D.J.; Fort, M.; Van Doorsselaere, J.; Nauwynck, H.J. Antigenic subtyping and epitopes’ competition analysis of porcine circovirus type 2 using monoclonal antibodies. Veter. Microbiol. 2012, 157, 13–22. [Google Scholar] [CrossRef]
- Kurtz, S.; Grau-Roma, L.; Cortey, M.; Fort, M.; Rodríguez, F.; Sibila, M.; Segalés, J.J. Pigs naturally exposed to porcine circovirus type 2 (PCV2) generate antibody responses capable to neutralise PCV2 isolates of different genotypes and geographic origins. Veter. Res. 2014, 45, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azmi, F.; Ahmad Fuaad, A.A.H.; Skwarczynski, M.; Toth, I. Recent progress in adjuvant discovery for peptide-based subunit vaccines. Hum. Vaccin. Immunother. 2014, 10, 778–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purcell, A.W.; McCluskey, J.; Rossjohn, J. More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. Drug Discov. 2007, 6, 404–414. [Google Scholar] [CrossRef]
- Vine, W.; Bowers, L.D. Cyclosporine: Structure, pharmacokinetics, and therapeutic drug monitoring. Crit. Rev. Clin. Lab. Sci. 1987, 25, 275–311. [Google Scholar] [CrossRef] [PubMed]
- Skwarczynski, M.; Toth, I. Peptide-based synthetic vaccines. Chem. Sci. 2016, 7, 842–854. [Google Scholar] [CrossRef] [Green Version]
- Coffman, R.L.; Sher, A.; Seder, R.A. Vaccine Adjuvants: Putting Innate Immunity to Work. Immunity 2010, 33, 492–503. [Google Scholar] [CrossRef] [Green Version]
- Duong, V.T.; Skwarczynski, M.; Toth, I. Towards the development of subunit vaccines against tuberculosis: The key role of adjuvant. Tuberculosis 2023, 139, 102307. [Google Scholar] [CrossRef]
- Firdaus, F.Z.; Skwarczynski, M.; Toth, I. Developments in Vaccine Adjuvants. Methods Mol. Biol. 2022, 2412, 145–178. [Google Scholar]
- Bosch, B.J.; van der Zee, R.; De Haan, C.A.; Rottier, P.J. The coronavirus spike protein is a class I virus fusion protein: Structural and functional characterization of the fusion core complex. J. Virol. 2003, 77, 8801–8811. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Khan, S.; Saleem, S.; Nizam-Uddin, N.; Mohammad, A.; Khan, T.; Ahmad, S.; Arshad, M.; Ali, S.S.; Suleman, M.; et al. Immunogenomics guided design of immunomodulatory multi-epitope subunit vaccine against the SARS-CoV-2 new variants, and its validation through in silico cloning and immune simulation. Comput. Biol. Med. 2021, 133, 104420. [Google Scholar] [CrossRef]
- Grela, P.; Koper, P.; Kutkowska, J.; Kwit, K.; Mazur, A.; Pejsak, Z.; Pochodyła, A.; Podgórska, K.; Rapak, A.; Skorupska, A.; et al. Vaccine for Porcine Circovirus Infection and Method for Obtaining it. Patent PL236294B1, 25 October 2018. [Google Scholar]
- Bachmann, M.F.; Jennings, G.T. Vaccine delivery: A matter of size, geometry, kinetics and molecular patterns. Nat. Rev. Immunol. 2010, 10, 787–796. [Google Scholar] [CrossRef]
- Shalash, A.O.; Becker, L.; Yang, J.; Giacomin, P.; Pearson, M.; Hussein, W.M.; Loukas, A.; Toth, I.; Skwarczynski, M. Development of a peptide vaccine against hookworm infection: Immunogenicity, efficacy, and immune correlates of protection. J. Allergy Clin. Immunol. 2022, 150, 157–169.e10. [Google Scholar] [CrossRef] [PubMed]
- Shalash, A.O.; Becker, L.; Yang, J.; Giacomin, P.; Pearson, M.; Hussein, W.M.; Loukas, A.; Skwarczynski, M.; Toth, I. Oral Peptide Vaccine against Hookworm Infection: Correlation of Antibody Titers with Protective Efficacy. Vaccines 2021, 9, 1034. [Google Scholar] [CrossRef]
- Yu, M.; Zeng, W.; Pagnon, J.; Walker, J.; Ghosh, S.; Wang, L.F.; Jackson, D.C. Identification of dominant epitopes of synthetic immunocontraceptive vaccines that induce antibodies in dogs. Vaccine 2005, 23, 4589–4597. [Google Scholar] [CrossRef]
- Azuar, A.; Jin, W.; Mukaida, S.; Hussein, W.; Toth, I.; Skwarczynski, M. Recent advances in the development of peptide vaccines and their delivery systems against Group A Streptococcus. Vaccines 2019, 7, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liechty, W.B.; Kryscio, D.R.; Slaughter, B.V.; Peppas, N.A. Polymers for drug delivery systems. Annu. Rev. Chem. Biomol. Eng. 2010, 1, 149–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nevagi, R.J.; Skwarczynski, M.; Toth, I. Polymers for subunit vaccine delivery. Eur. Polym. J. 2019, 114, 397–410. [Google Scholar] [CrossRef]
- Adams, J.R.; Haughney, S.L.; Mallapragada, S.K. Effective polymer adjuvants for sustained delivery of protein subunit vaccines. Acta Biomater. 2015, 14, 104–114. [Google Scholar] [CrossRef]
- Lone, N.A.; Spackman, E.; Kapczynski, D. Immunologic evaluation of 10 different adjuvants for use in vaccines for chickens against highly pathogenic avian influenza virus. Vaccine 2017, 35, 3401–3408. [Google Scholar] [CrossRef]
- Park, M.-E.; Lee, S.-Y.; Kim, R.-H.; Ko, M.-K.; Lee, K.-N.; Kim, S.-M.; Kim, B.-K.; Lee, J.-S.; Kim, B.; Park, J.-H. Enhanced immune responses of foot-and-mouth disease vaccine using new oil/gel adjuvant mixtures in pigs and goats. Vaccine 2014, 32, 5221–5227. [Google Scholar] [CrossRef]
- Gong, X.; Chen, Q.; Ferguson-Noel, N.; Stipkovits, L.; Szathmary, S.; Liu, Y.; Zheng, F. Evaluation of protective efficacy of inactivated Mycoplasma synoviae vaccine with different adjuvants. Vet. Immunol. Immunopathol. 2020, 220, 109995. [Google Scholar] [CrossRef]
- Yang, Y.; Xing, R.; Liu, S.; Qin, Y.; Li, K.; Yu, H.; Li, P. Chitosan, hydroxypropyltrimethyl ammonium chloride chitosan and sulfated chitosan nanoparticles as adjuvants for inactivated Newcastle disease vaccine. Carbohydr. Polym. 2020, 229, 115423. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Zaman, M.; Urbani, C.N.; Lin, I.C.; Jia, Z.; Batzloff, M.R.; Good, M.F.; Monteiro, M.J.; Toth, I. Polyacrylate dendrimer nanoparticles: A self-adjuvanting vaccine delivery system. Angew. Chem. 2010, 122, 5878–5881. [Google Scholar] [CrossRef]
- Skwarczynski, M.; Zhao, G.; Boer Jennifer, C.; Ozberk, V.; Azuar, A.; Cruz Jazmina, G.; Giddam Ashwini, K.; Khalil Zeinab, G.; Pandey, M.; Shibu Mohini, A.; et al. Poly(amino acids) as a potent self-adjuvanting delivery system for peptide-based nanovaccines. Sci. Adv. 2020, 6, eaax2285. [Google Scholar] [CrossRef] [Green Version]
- Azuar, A.; Li, Z.; Shibu, M.A.; Zhao, L.; Luo, Y.; Shalash, A.O.; Khalil, Z.G.; Capon, R.J.; Hussein, W.M.; Toth, I.; et al. Poly(hydrophobic amino acid)-Based Self-Adjuvanting Nanoparticles for Group A Streptococcus Vaccine Delivery. J. Med. Chem. 2021, 64, 2648–2658. [Google Scholar] [CrossRef] [PubMed]
- Azuar, A.; Shibu, M.A.; Adilbish, N.; Marasini, N.; Hung, H.; Yang, J.; Luo, Y.; Khalil, Z.G.; Capon, R.J.; Hussein, W.M.; et al. Poly(hydrophobic amino acid) Conjugates for the Delivery of Multiepitope Vaccine against Group a Streptococcus. Bioconjugate Chem. 2021, 32, 2307–2317. [Google Scholar] [CrossRef]
- Bartlett, S.; Skwarczynski, M.; Xie, X.; Toth, I.; Loukas, A.; Eichenberger, R. Development of natural and unnatural amino acid delivery systems against hookworm infection. Precis. Nanomed. 2020, 3, 471–482. [Google Scholar] [CrossRef] [Green Version]
- Bozzuto, G.; Molinari, A. Liposomes as nanomedical devices. Int. J. Nanomed. 2015, 10, 975–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Çağdaş, M.; Sezer, A.D.; Bucak, S. Liposomes as potential drug carrier systems for drug delivery. Appl. Nanotechnol. Drug Deliv. 2014, 1, 1–50. [Google Scholar] [CrossRef] [Green Version]
- De Serrano, L.O.; Burkhart, D.J. Liposomal vaccine formulations as prophylactic agents: Design considerations for modern vaccines. J. Nanobiotechnol. 2017, 15, 83. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Azuar, A.; Toth, I.; Skwarczynski, M. Liposomes for the delivery of lipopeptide vaccines. Vaccine Des. 2021, 2412, 295–307. [Google Scholar] [CrossRef]
- Azuar, A.; Madge, H.Y.R.; Boer, J.C.; Gonzalez Cruz, J.L.; Wang, J.; Khalil, Z.G.; Deceneux, C.; Goodchild, G.; Yang, J.; Koirala, P.; et al. Poly(hydrophobic Amino Acids) and Liposomes for Delivery of Vaccine against Group A Streptococcus. Vaccines 2022, 10, 1212. [Google Scholar] [CrossRef]
- Wijburg, O.L.; Heemskerk, M.H.; Boog, C.J.; Van Rooijen, N. Role of spleen macrophages in innate and acquired immune responses against mouse hepatitis virus strain A59. Immunology 1997, 92, 252–258. [Google Scholar] [CrossRef] [Green Version]
- Gu, W.; Bobrin, V.A.; Chen, S.-P.R.; Wang, Z.; Schoning, J.P.; Gu, Y.; Chen, W.; Chen, M.; Jia, Z.; Monteiro, M.J. Biodistribution of PNIPAM-Coated Nanostructures Synthesized by the TDMT Method. Biomacromolecules 2019, 20, 625–634. [Google Scholar] [CrossRef]
- Koirala, P.; Chen, S.-P.R.; Boer, J.C.; Khalil, Z.G.; Deceneux, C.; Goodchild, G.; Lu, L.; Faruck, M.O.; Shalash, A.O.; Bashiri, S.; et al. Polymeric Nanoparticles as a Self-Adjuvanting Peptide Vaccine Delivery System: The Role of Shape. Adv. Funct. Mater. 2023, 2209304. [Google Scholar] [CrossRef]
- Waleed, M.; Hussein, M.S.; Istvan, T. Peptide Synthesis, Methods and Protocols, 1st ed.; Humana: New York, NY, USA, 2021. [Google Scholar]
- Chandrudu, S.; Bartlett, S.; Khalil, Z.G.; Jia, Z.; Hussein, W.M.; Capon, R.J.; Batzloff, M.R.; Good, M.F.; Monteiro, M.J.; Skwarczynski, M. Linear and branched polyacrylates as a delivery platform for peptide-based vaccines. Ther. Deliv. 2016, 7, 601–609. [Google Scholar] [CrossRef] [PubMed]
- Fort, M.; Olvera, A.; Sibila, M.; Segalés, J.; Mateu, E. Detection of neutralizing antibodies in postweaning multisystemic wasting syndrome (PMWS)-affected and non-PMWS-affected pigs. Vet. Microbiol. 2007, 125, 244–255. [Google Scholar] [CrossRef] [Green Version]
- Petrovsky, N.; Aguilar, J.C. Vaccine adjuvants: Current state and future trends. Immunol. Cell Biol. 2004, 82, 488–496. [Google Scholar] [CrossRef]
- Champion, J.A.; Mitragotri, S. Shape induced inhibition of phagocytosis of polymer particles. Pharm. Res. 2009, 26, 244–249. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Vedi, S.; Kunimoto, D.Y.; Agrawal, B.; Kumar, R. Novel lipopeptides of ESAT-6 induce strong protective immunity against Mycobacterium tuberculosis: Routes of immunization and TLR agonists critically impact vaccine’s efficacy. Vaccine 2016, 34, 5677–5688. [Google Scholar] [CrossRef]
- Sun, B.; Xia, T. Nanomaterial-based vaccine adjuvants. J. Mater. Chem. B 2016, 4, 5496–5509. [Google Scholar] [CrossRef] [PubMed]
- Fifis, T.; Gamvrellis, A.; Crimeen-Irwin, B.; Pietersz, G.A.; Li, J.; Mottram, P.L.; McKenzie, I.F.; Plebanski, M. Size-dependent immunogenicity: Therapeutic and protective properties of nano-vaccines against tumors. J. Immunol. 2004, 173, 3148–3154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zinkhan, S.; Ogrina, A.; Balke, I.; Reseviča, G.; Zeltins, A.; de Brot, S.; Lipp, C.; Chang, X.; Zha, L.; Vogel, M. The impact of size on particle drainage dynamics and antibody response. J. Control. Release 2021, 331, 296–308. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liu, Z.H.; Li, Y.X.; Xu, H.L.; Fang, W.H.; He, F. Targeted Delivery of Nanovaccine to Dendritic Cells via DC-Binding Peptides Induces Potent Antiviral Immunity in vivo. Int. J. Nanomed. 2022, 17, 1593–1608. [Google Scholar] [CrossRef] [PubMed]
- Rashidzadeh, H.; Danafar, H.; Rahimi, H.; Mozafari, F.; Salehiabar, M.; Rahmati, M.A.; Rahamooz-Haghighi, S.; Mousazadeh, N.; Mohammadi, A.; Ertas, Y.N.; et al. Nanotechnology against the novel coronavirus (severe acute respiratory syndrome coronavirus 2): Diagnosis, treatment, therapy and future perspectives. Nanomedicine 2021, 16, 497–516. [Google Scholar] [CrossRef]
- Ding, P.; Jin, Q.; Chen, X.; Yang, S.; Guo, J.; Xing, G.; Deng, R.; Wang, A.; Zhang, G. Nanovaccine Confers Dual Protection Against Influenza A Virus And Porcine Circovirus Type 2. Int. J. Nanomed. 2019, 14, 7533–7548. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Anselmo, A.C.; Banerjee, A.; Zakrewsky, M.; Mitragotri, S. Shape and size-dependent immune response to antigen-carrying nanoparticles. J. Control. Release 2015, 220, 141–148. [Google Scholar] [CrossRef]
- Nevagi, R.J.; Dai, W.; Khalil, Z.G.; Hussein, W.M.; Capon, R.J.; Skwarczynski, M.; Toth, I. Structure-activity relationship of group A streptococcus lipopeptide vaccine candidates in trimethyl chitosan-based self-adjuvanting delivery system. Eur. J. Med. Chem. 2019, 179, 100–108. [Google Scholar] [CrossRef]
- Azuar, A.; Zhao, L.; Hei, T.T.; Nevagi, R.J.; Bartlett, S.; Hussein, W.M.; Khalil, Z.G.; Capon, R.J.; Toth, I.; Skwarczynski, M. Cholic acid-based delivery system for vaccine candidates against group A streptococcus. ACS Med. Chem. Lett. 2019, 10, 1253–1259. [Google Scholar] [CrossRef] [PubMed]
- Hotaling, N.A.; Cummings, R.D.; Ratner, D.M.; Babensee, J.E. Molecular factors in dendritic cell responses to adsorbed glycoconjugates. Biomaterials 2014, 35, 5862–5874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad Fuaad, A.A.; Jia, Z.; Zaman, M.; Hartas, J.; Ziora, Z.M.; Lin, I.C.; Moyle, P.M.; Batzloff, M.R.; Good, M.F.; Monteiro, M.J.; et al. Polymer-peptide hybrids as a highly immunogenic single-dose nanovaccine. Nanomedicine 2014, 9, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Martins, K.A.; Bavari, S.; Salazar, A.M. Vaccine adjuvant uses of poly-IC and derivatives. Expert Rev. Vaccines 2015, 14, 447–459. [Google Scholar] [CrossRef]



| Vaccine | Antigen | Adjuvant | Administration |
|---|---|---|---|
| Circovac® | Inactivated PCV2a | Light paraffin oil | Single dose, intramuscular (IM) for piglets older than 3 weeks or healthy female pigs of breeding age; or two doses, prior to breeding for gilts and sows. |
| Ingelvac CircoFlex® | Capsid PCV2a | Cross-linked carbomer-polymer | Single dose, IM |
| Circumvent® | Capsid PCV2a | D1-α-tocopherol + liquid paraffin | Two doses, IM (3 weeks old) |
| Porcillis® PCV | Capsid PCV2a | D1-α-tocopherol + liquid paraffin | Two doses, IM (3 days old) or Single dose, IM (3 weeks old) |
| Fostera™ PCV (Suvaxyn® PCV2 one dose™) | Inactivated chimaeric PCV1/2a | Sulpholipo-cyclodextrin in squalane | Single dose, IM (3 weeks old) |
| Vaccine Candidate | Antigen | Adjuvanting Moiety | Size (nm) | PDI | Zeta Potential (mV) |
|---|---|---|---|---|---|
| V1 | Cap41, Cap67, Cap121, P25 | PMA | 280 ± 8 | 0.23 ± 0.01 | 43.7 ± 0.7 |
| V2 | Cap41, Cap67, Cap121, P25 | Leu10 | 86 ± 19 | 0.57 ± 0.20 | 34.0 ± 0.1 |
| 363 ± 54 | |||||
| V3 | Cap41, Cap67, Cap121, P25 | Leu10, liposome | 206 ± 3 | 0.26 ± 0.01 | 25.0 ± 0.8 |
| V4 | Cap41, Cap67, Cap121, P25 | rods | 158 ± 15 | 0.42 ± 0.07 | 11.9 ± 0.7 |
| 503 ± 55 | |||||
| 5200 ± 180 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duong, V.T.; Koirala, P.; Chen, S.-P.R.; Monteiro, M.J.; Skwarczynski, M.; Toth, I. Multiepitope Subunit Peptide-Based Nanovaccine against Porcine Circovirus Type 2 (PCV2) Elicited High Antibody Titers in Vaccinated Mice. Molecules 2023, 28, 2248. https://doi.org/10.3390/molecules28052248
Duong VT, Koirala P, Chen S-PR, Monteiro MJ, Skwarczynski M, Toth I. Multiepitope Subunit Peptide-Based Nanovaccine against Porcine Circovirus Type 2 (PCV2) Elicited High Antibody Titers in Vaccinated Mice. Molecules. 2023; 28(5):2248. https://doi.org/10.3390/molecules28052248
Chicago/Turabian StyleDuong, Viet Tram, Prashamsa Koirala, Sung-Po R. Chen, Michael J. Monteiro, Mariusz Skwarczynski, and Istvan Toth. 2023. "Multiepitope Subunit Peptide-Based Nanovaccine against Porcine Circovirus Type 2 (PCV2) Elicited High Antibody Titers in Vaccinated Mice" Molecules 28, no. 5: 2248. https://doi.org/10.3390/molecules28052248
APA StyleDuong, V. T., Koirala, P., Chen, S. -P. R., Monteiro, M. J., Skwarczynski, M., & Toth, I. (2023). Multiepitope Subunit Peptide-Based Nanovaccine against Porcine Circovirus Type 2 (PCV2) Elicited High Antibody Titers in Vaccinated Mice. Molecules, 28(5), 2248. https://doi.org/10.3390/molecules28052248