Quinoline-Malononitrile-Based Aggregation-Induced Emission Probe for Monoamine Oxidase Detection in Living Cells
Abstract
:1. Introduction
2. Results and Discussions
2.1. Synthesis

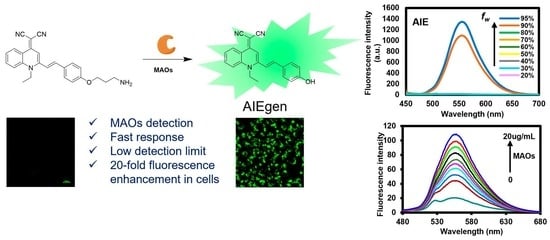
2.2. Photophysical Properties
2.2.1. Fluorescence Responses of QM-NH2 to MAOs
2.2.2. HPLC Analysis of the Enzymatic Reaction
2.2.3. Selectivity of QM-NH2 toward MAOs
2.2.4. Interactions of QM-NH2 and MAOs Visualized by Molecular Docking
2.3. Cellular Assays
3. Materials and Instruments
3.1. UV–vis and Fluorescence Spectroscopic Methods
3.2. MAO Assays
3.3. Cell Culture
3.4. Cell Viability Assay
3.5. Time-Dependent Internalization
3.6. Time Course Live Cell Imaging
3.7. Cell Fixation Assay
3.8. Inhibition Assay
3.9. Statistical Analysis
3.10. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Shih, J.C.; Chen, K.; Ridd, M.J. Role of MAO A and B in neurotransmitter metabolism and behavior. Pol. J. Pharmacol. 1999, 51, 25–29. [Google Scholar] [PubMed]
- Shih, J.C.; Chen, K.; Ridd, M.J. Monoamine oxidase: From genes to behavior. Annu. Rev. Neurosci. 1999, 22, 197–217. [Google Scholar] [CrossRef] [PubMed]
- Edmondson, D.E.; Mattevi, A.; Binda, C.; Li, M.; Hubalek, F. Structure and mechanism of monoamine oxidase. Curr. Med. Chem. 2004, 11, 1983–1993. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Li, W.; Chen, H.; Mo, Q.; Li, J.; Zhao, S.; Hou, C.; Qin, J.; Su, G. Inhibitor structure-guided design and synthesis of near-infrared fluorescent probes for monoamine oxidase A (MAO-A) and its application in living cells and in vivo. Chem. Commun. 2019, 55, 2477–2480. [Google Scholar] [CrossRef] [PubMed]
- Mattson, M.P. Apoptosis in neurodegenerative disorders. Nat. Rev. Mol. Cell Biol. 2000, 1, 120–129. [Google Scholar] [CrossRef]
- Maragos, W.F.; Young, K.L.; Altman, C.S.; Pocernich, C.B.; Drake, J.; Butterfield, D.A.; Seif, I.; Holschneider, D.P.; Chen, K.; Shih, J.C. Striatal damage and oxidative stress induced by the mitochondrial toxin malonate are reduced in clorgyline-treated rats and MAO-A deficient mice. Neurochem. Res. 2004, 29, 741–746. [Google Scholar] [CrossRef]
- Geha, R.M.; Rebrin, I.; Chen, K.; Shih, J.C. Substrate and inhibitor specificities for human monoamine oxidase A and B are influenced by a single amino acid. J. Biol. Chem. 2001, 276, 9877–9882. [Google Scholar] [CrossRef]
- Youdim, M.B.; Buccafusco, J.J. CNS Targets for multi-functional drugs in the treatment of Alzheimer’s and Parkinson’s diseases. J. Neural. Transm. 2005, 112, 519–537. [Google Scholar] [CrossRef]
- Wang, C.C.; Billett, E.; Borchert, A.; Kuhn, H.; Ufer, C. Monoamine oxidases in development. Cell. Mol. Life Sci. 2013, 70, 599–630. [Google Scholar] [CrossRef]
- Murphy, D.L.; Lipper, S.; Slater, S.; Shiling, D. Selectivity of clorgyline and pargyline as inhibitors of monoamine oxidases A and B in vivo in man. Psychopharmacology 1979, 62, 129–132. [Google Scholar] [CrossRef]
- McDermott, R.; Tingley, D.; Cowden, J.; Frazzetto, G.; Johnson, D.D. Monoamine oxidase A gene (MAOA) predicts behavioral aggression following provocation. Proc. Natl. Acad. Sci. USA 2009, 106, 2118–2123. [Google Scholar] [CrossRef]
- Guo, Z.; Yan, C.; Zhu, W.H. High-Performance Quinoline-Malononitrile Core as a Building Block for the Diversity-Oriented Synthesis of AIEgens. Angew. Chem. Int. Ed. 2020, 59, 9812–9825. [Google Scholar] [CrossRef]
- Shi, C.; Guo, Z.; Yan, Y.; Zhu, S.; Xie, Y.; Zhao, Y.S.; Zhu, W.; Tian, H. Self-assembly solid-state enhanced red emission of quinolinemalononitrile: Optical waveguides and stimuli response. ACS Appl. Mater. Interfaces 2013, 5, 192–198. [Google Scholar] [CrossRef]
- Guo, Z.; Zhu, W.; Tian, H. Dicyanomethylene-4H-pyran chromophores for OLED emitters, logic gates and optical chemosensors. Chem. Commun. 2012, 48, 6073–6084. [Google Scholar] [CrossRef]
- Díaz, I.; Kokkoli, E.; Terasaki, O.; Tsapatsis, M. Surface structure of zeolite (MFI) crystals. Chem. Mater. 2004, 16, 5226–5232. [Google Scholar] [CrossRef]
- Chen, C.-T. Evolution of red organic light-emitting diodes: Materials and devices. Chem. Mater. 2004, 16, 4389–4400. [Google Scholar] [CrossRef]
- Wu, X.; Li, Z.; Chen, X.-X.; Fossey, J.S.; James, T.D.; Jiang, Y.-B. Selective sensing of saccharides using simple boronic acids and their aggregates. Chem. Soc. Rev. 2013, 42, 8032–8048. [Google Scholar] [CrossRef]
- Chan, J.; Dodani, S.C.; Chang, C.J. Reaction-based small-molecule fluorescent probes for chemoselective bioimaging. Nat. Chem. 2012, 4, 973–984. [Google Scholar] [CrossRef]
- Myochin, T.; Hanaoka, K.; Komatsu, T.; Terai, T.; Nagano, T. Design strategy for a near-infrared fluorescence probe for matrix metalloproteinase utilizing highly cell permeable boron dipyrromethene. J. Am. Chem. Soc. 2012, 134, 13730–13737. [Google Scholar] [CrossRef]
- Guo, Z.; Shao, A.; Zhu, W.-H. Long wavelength AIEgen of quinoline-malononitrile. J. Mater. Chem. C 2016, 4, 2640–2646. [Google Scholar] [CrossRef]
- Luo, J.; Xie, Z.; Lam, J.W.; Cheng, L.; Chen, H.; Qiu, C.; Kwok, H.S.; Zhan, X.; Liu, Y.; Zhu, D.; et al. Aggregation-induced emission of 1-methyl-1,2,3,4,5-pentaphenylsilole. Chem. Commun. 2001, 18, 1740–1741. [Google Scholar] [CrossRef] [PubMed]
- Pramanik, S.; Bhalla, V.; Kumar, M. Hexaphenylbenzene-based fluorescent aggregates for ratiometric detection of cyanide ions at nanomolar level: Set-reset memorized sequential logic device. ACS Appl. Mater. Interfaces 2014, 6, 5930–5939. [Google Scholar] [CrossRef]
- An, B.K.; Kwon, S.K.; Jung, S.D.; Park, S.Y. Enhanced emission and its switching in fluorescent organic nanoparticles. J. Am. Chem. Soc. 2002, 124, 14410–14415. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Liu, G.; Qiao, M.; Li, Y.; Yi, X. Biosensors for the Detection of Enzymes Based on Aggregation-Induced Emission. Biosensors 2022, 12, 953. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yee, D.J.; Gubernator, N.G.; Sames, D. Design of optical switches as metabolic indicators: New fluorogenic probes for monoamine oxidases (MAO A and B). J. Am. Chem. Soc. 2005, 127, 4544–4545. [Google Scholar] [CrossRef]
- Zhou, W.; Valley, M.P.; Shultz, J.; Hawkins, E.M.; Bernad, L.; Good, T.; Good, D.; Riss, T.L.; Klaubert, D.H.; Wood, K.V. New bioluminogenic substrates for monoamine oxidase assays. J. Am. Chem. Soc. 2006, 128, 3122–3123. [Google Scholar] [CrossRef]
- Kim, D.; Sambasivan, S.; Nam, H.; Kim, K.H.; Kim, J.Y.; Joo, T.; Lee, K.H.; Kim, K.T.; Ahn, K.H. Reaction-based two-photon probes for in vitro analysis and cellular imaging of monoamine oxidase activity. Chem. Commun. 2012, 48, 6833–6835. [Google Scholar] [CrossRef]
- Loo, L.S.; Tang, N.; Al-Haddawi, M.; Dawe, G.S.; Hong, W. A role for sorting nexin 27 in AMPA receptor trafficking. Nat. Commun. 2014, 5, 3176. [Google Scholar] [CrossRef]
- Li, L.; Zhang, C.W.; Ge, J.; Qian, L.; Chai, B.H.; Zhu, Q.; Lee, J.S.; Lim, K.L.; Yao, S.Q. A Small-Molecule Probe for Selective Profiling and Imaging of Monoamine Oxidase B Activities in Models of Parkinson’s Disease. Angew. Chem. Int. Ed. 2015, 54, 10821–10825. [Google Scholar] [CrossRef]
- Li, L.L.; Li, K.; Liu, Y.H.; Xu, H.R.; Yu, X.Q. Red emission fluorescent probes for visualization of monoamine oxidase in living cells. Sci. Rep. 2016, 6, 31217. [Google Scholar] [CrossRef]
- Han, X.; Yang, X.; Zhang, Y.; Li, Z.; Cao, W.; Zhang, D.; Ye, Y. A novel activatable AIEgen fluorescent probe for peroxynitrite detection and its application in EC1 cells. Sens. Actuators B Chem. 2020, 321, 128510. [Google Scholar] [CrossRef]
- Wang, M.; Yang, N.; Guo, Z.; Gu, K.; Shao, A.; Zhu, W.; Xu, Y.; Wang, J.; Prud’homme, R.K.; Guo, X. Facile Preparation of AIE-Active Fluorescent Nanoparticles through Flash Nanoprecipitation. Ind. Eng. Chem. Res. 2015, 54, 4683–4688. [Google Scholar] [CrossRef]
- Li, L.; Zhang, C.W.; Chen, G.Y.; Zhu, B.; Chai, C.; Xu, Q.H.; Tan, E.K.; Zhu, Q.; Lim, K.L.; Yao, S.Q. A sensitive two-photon probe to selectively detect monoamine oxidase B activity in Parkinson’s disease models. Nat. Commun. 2014, 5, 3276. [Google Scholar] [CrossRef]
- Van Meerloo, J.; Kaspers, G.J.; Cloos, J. Cell sensitivity assays: The MTT assay. Methods Mol. Biol. 2011, 731, 237–245. [Google Scholar]
- Naoi, M.; Maruyama, W.; Akao, Y.; Yi, H.; Yamaoka, Y. Involvement of type A monoamine oxidase in neurodegeneration: Regulation of mitochondrial signaling leading to cell death or neuroprotection. In Oxidative Stress and Neuroprotection; Springer: Berlin, Germany, 2006; pp. 67–77. [Google Scholar]
- Shih, J.C.; Chen, K. Regulation of MAO-A and MAO-B gene expression. Curr. Med. Chem. 2004, 11, 1995–2005. [Google Scholar] [CrossRef]
- Aljanabi, R.; Alsous, L.; Sabbah, D.A.; Gul, H.I.; Gul, M.; Bardaweel, S.K. Monoamine Oxidase (MAO) as a Potential Target for Anticancer Drug Design and Development. Molecules 2021, 26, 6019. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Giacovazzo, C.; Capitelli, F.; Cuocci, C.; Ianigro, M. Direct methods and applications to electron crystallography. In Microscopy, Spectroscopy, Holography and Crystallography with Electrons; Hawkes, P.W., Merli, P.G., Calestani, G., Vittori-Antisari, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2002; Volume 123, pp. 291–310. [Google Scholar]







Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duangkamol, C.; Wangngae, S.; Wet-osot, S.; Khaikate, O.; Chansaenpak, K.; Lai, R.-Y.; Kamkaew, A. Quinoline-Malononitrile-Based Aggregation-Induced Emission Probe for Monoamine Oxidase Detection in Living Cells. Molecules 2023, 28, 2655. https://doi.org/10.3390/molecules28062655
Duangkamol C, Wangngae S, Wet-osot S, Khaikate O, Chansaenpak K, Lai R-Y, Kamkaew A. Quinoline-Malononitrile-Based Aggregation-Induced Emission Probe for Monoamine Oxidase Detection in Living Cells. Molecules. 2023; 28(6):2655. https://doi.org/10.3390/molecules28062655
Chicago/Turabian StyleDuangkamol, Chuthamat, Sirilak Wangngae, Sirawit Wet-osot, Onnicha Khaikate, Kantapat Chansaenpak, Rung-Yi Lai, and Anyanee Kamkaew. 2023. "Quinoline-Malononitrile-Based Aggregation-Induced Emission Probe for Monoamine Oxidase Detection in Living Cells" Molecules 28, no. 6: 2655. https://doi.org/10.3390/molecules28062655
APA StyleDuangkamol, C., Wangngae, S., Wet-osot, S., Khaikate, O., Chansaenpak, K., Lai, R. -Y., & Kamkaew, A. (2023). Quinoline-Malononitrile-Based Aggregation-Induced Emission Probe for Monoamine Oxidase Detection in Living Cells. Molecules, 28(6), 2655. https://doi.org/10.3390/molecules28062655