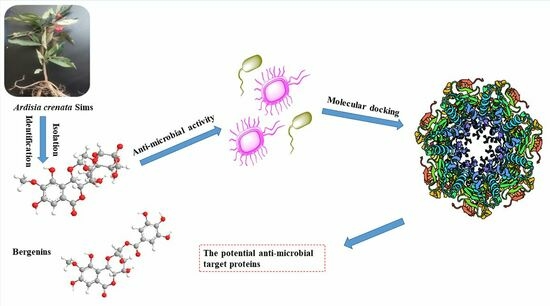
Discovery of Potential Anti-Microbial Molecules and Spectrum Correlation Effect of Ardisia crenata Sims via High-Performance Liquid Chromatography Fingerprints and Molecular Docking
Abstract
:1. Introduction
2. Results and Discussion
2.1. Anti-Microbial Activity Evaluation of A. crenata Sims
2.2. Structural Identification of Known Compounds from A. crenata Sims
2.3. Analysis of HPLC Fingerprint
2.4. PCA and HCA Analysis of 16 Batches of A. crenata Sims
2.5. Effect of Compounds 1–5 on Anti-Microbial Activities
2.6. Spectrum–Effect Relationship
2.7. Quantitative Analysis of Anti-Microbial Ingredients in A. crenata Sims
2.8. Molecular Docking of Active Compounds on Key Target Proteins of Bacteria
2.9. Molecular Docking of Active Compounds on Key Target Proteins of Fungi
3. Materials and Methods
3.1. Instruments and Chemicals
3.2. Plant Materials and Test Strains
3.3. Extraction and Isolation of Roots from A. crenata Sims
3.4. Sample Preparation
3.5. Chromatographic Conditions
3.6. Validation of Methodology
3.7. Analysis of HPLC Fingerprint
3.8. Anti-Bacterial Activity Evaluation
3.9. Spectrum-Effect Relationship
3.9.1. Gray Relational Analysis (GRA)
3.9.2. Partial Least Squares Regression (PLSR)
3.10. Molecular Docking Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Schmidt, T.; Kock, M.M.; Ehlers, M.M. Molecular characterization of Staphylococcus aureus isolated from bovine mastitis and close human contacts in South African dairy herds: Genetic diversity and inter-species host transmission. Front. Microbiol. 2017, 8, 511. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Tian, L.; Tan, B.; Shen, Z.H.; Xiao, M.W.; Wu, S.; Meng, X.R.; Wu, X.; Wang, X.Y. Update: Innate Lymphoid Cells in Inflammatory Bowel Disease. Dig. Dis. Sci. 2022, 67, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Jing, S.S.; Kong, X.R.; Wang, L.; Wang, H.M.; Feng, J.X.; Wei, L.; Meng, Y.; Liu, C.; Chang, X.R.; Qu, Y.S.; et al. Quercetin reduces the virulence of S. aureus by targeting ClpP to protect mice from MRSA-induced lethal pneumonia. Microbiol. Spectr. 2022, 10, e02340-21. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.Y.; Zhang, T.; Wang, P.Y.; Pan, Y.H.; Li, J.H.; Chen, W.Z.; Zhang, M.; Ji, Q.J.; Wu, W.J.; Lan, L.F.; et al. Anti-infective therapy using species-specific activators of Staphylococcus aureus ClpP. Nat. Commun. 2022, 13, 6909. [Google Scholar] [CrossRef] [PubMed]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.G.; Xiao, W.; Zhou, C.M.; Pu, Q.Q.; Deng, X.; Lan, L.F.; Liang, H.H.; Song, X.R.; Wu, M. Pseudomonas aeruginosa: Pathogenesis, virulence factors, antibiotic resistance, interaction with host, technology advances and emerging therapeutics. Signal Transduct. Target. Ther. 2022, 7, 199. [Google Scholar] [CrossRef]
- Casilag, F.; Lorenz, A.; Krueger, J.; Klawonn, F.; Weiss, S.; Häussler, S. The LasB elastase of Pseudomonas aeruginosa acts in concert with alkaline protease AprA to prevent flagellin-mediated immune recognition. Infect. Immun. 2016, 84, 162–171. [Google Scholar] [CrossRef]
- Horcajada, J.P.; Montero, M.; Oliver, A.; Sorlí, L.; Luque, S.; Gómez-Zorrilla, S.; Benito, N.; Grau, S. Epidemiology and treatment of multidrug-resistant and extensively drug-resistant Pseudomonas aeruginosa infections. Clin. Microbiol. Rev. 2019, 32, e00031-19. [Google Scholar] [CrossRef]
- Uddin, T.M.; Chakraborty, A.J.; Khusro, A.; Zidan, B.R.M.; Mitra, S.; Emran, T.B.; Koirala, N. Antibiotic resistance in microbes: History, mechanisms, therapeutic strategies and future prospects. J. Infect. Public Health 2021, 14, 1750–1766. [Google Scholar] [CrossRef]
- Durand, G.A.; Raoult, D.; Dubourg, G. Antibiotic discovery: History, methods and perspectives. Int. J. Antimicrob. Agents 2019, 53, 371–382. [Google Scholar] [CrossRef]
- Sun, S.J.; Deng, P.; Peng, C.E.; Ji, H.Y.; Mao, L.F.; Peng, L.Z. Extraction, structure and immunoregulatory activity of low molecular weight polysaccharide from Dendrobium officinale. Polymers 2022, 14, 2899. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ma, J.; Li, Y.; Han, Q.; Yin, Z.; Zhou, M.; Luo, M.Y.; Chen, J.Y.; Xia, S.T. Effects of essential oil extracted from Artemisia argyi leaf on lipid metabolism and gut microbiota in high-fat diet-fed mice. Front. Nutr. 2022, 9, 1024722. [Google Scholar] [CrossRef]
- Ibrahim, M.; Riaz, M.; Ali, A.; Shaheen, M.; ur Rahman, S.; Aziz, R.; Alamri, A.S.; Alhomrani, M.; Dablool, A.S.; Alghamdi, S.; et al. Evaluating the total phenolic, protein contents, antioxidant and pharmacological effects of Cynodon dactylon extracts against Escherichia coli and Staphylococcus aureus. Pol. J. Chem. Technol. 2023, 25, 110–119. [Google Scholar] [CrossRef]
- Jiang, Q.J.; Yang, F.; Yang, A.D.; Wu, Y.; Lu, W.T.; Lei, S.Y.; Wu, X.L. Research progress of bacteriostatic mechanism of traditional Chinese medicine based on chemical components. Chin. J. Antibiot. 2023, 43, 855–861. [Google Scholar]
- Liu, F.; Peng, J.; Feng, Y.; Ma, Y.; Ren, Y.; Sun, P.; Zhao, Y.X.; Liu, S.; Wu, F.M.; Xie, J. An ethnobotanical study on the medicinal herb practices of the gelao ethnic minority in North Guizhou, China: An exploration of traditional knowledge. Front. Pharmacol. 2023, 14, 1217599. [Google Scholar] [CrossRef]
- Tao, H.; Zhou, Y.; Yin, X.; Wei, X.; Zhou, Y. Two New Phenolic Glycosides with Lactone Structural Units from Leaves of Ardisia crenata Sims with Antibacterial and Anti-Inflammatory Activities. Molecules 2022, 27, 4903. [Google Scholar] [CrossRef]
- Podolak, I.; Mynarski, A.; Wróbel, D.; Grabowska, K.; Galanty, A. Bioactive benzoquinones content variability in red-berry and white-berry varieties of Ardisia crenata Sims and assessment of cytotoxic activity. Nat. Prod. Res. 2021, 35, 157–161. [Google Scholar] [CrossRef]
- Tian, Z.H.; He, Y.; Luo, H.M.; Huang, Y.Q. Antibacterial and anti-inflammatory effects of Ardisia crenata Sims. Northwest Pharm. J. 1998, 13, 109–110. [Google Scholar]
- Ye, Q.; Chen, J.P.; Ling, Y.; Liu, Y.M.; Liu, Y.; Gai, X.H.; Tian, C.W.; Cheng, C.Q. Research progress on chemical constituents and pharmacological effects of Ardisiae Crenatae Radi. Chin. Tradit. Herb. Drugs 2022, 53, 2851–2860. [Google Scholar]
- Ding, S.; Tang, C.P.; Wu, C.Q.; Dong, Y.S.; Dong, X.; Wu, C.B.; Gu, C.B.; Fan, X.B.; Wang, Q.J.; Jiang, T. Toxicity study of thirteen⁃week repeated dose of Kaihoujian spray (child type) in juvenile rats. Mil. Med. 2021, 45, 516–525. [Google Scholar]
- Song, N.N.; Yang, L.M.; Zhang, M.J.; An, R.F.; Liu, W.; Huang, X.F. Triterpenoid saponins and phenylpropanoid glycoside from the roots of Ardisia crenata and their cytotoxic activities. Chin. J. Nat. Med. 2021, 19, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Shi, H.; Ding, J.X.; Feng, T.T.; Liu, X.W.; Liu, C.; Zhou, Y. Analysis of chemical constituents asflavonoids and coumarins in Radix Ardisiae from different sources. China Pharm. 2021, 32, 443–452. [Google Scholar]
- Salimo, Z.M.; Yakubu, M.N.; da Silva, E.L.; de Almeida, A.C.G.; Chaves, Y.O.; Costa, E.V.; da Silva, F.M.A.; Tavares, J.F.; Monteiro, W.M.; de Melo, G.C.; et al. Chemistry and pharmacology of bergenin or its derivatives: Apromising molecule. Biomolecules 2023, 13, 403. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Yu, S.; Duan, L.; Meng, S.; Ding, S.; Dong, T. New 1, 8-naphthalimide-based colorimetric fluorescent probe for specific detection of hydrazine and its multi-functional applications. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2024, 305, 123450. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, H.A.; Ahmad, S.; Gao, L.; Ismail, S.; Wang, Z.; El-Baz, A.; Ni, S.Q. Multi-omics analysis revealed the selective enrichment of partial denitrifying bacteria for the stable coupling of partial-denitrification and anammox process under the influence of low strength magnetic field. Water Res. 2023, 245, 120619. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Dong, Y.; Chen, X.; Wang, Z.; Cui, Z.; Ni, S.Q. Using sulfide as nitrite oxidizing bacteria inhibitor for the successful coupling of partial nitrification-anammox and sulfur autotrophic denitrification in one reactor. Chem. Eng. J. 2023, 475, 146286. [Google Scholar] [CrossRef]
- Serrano, L.; Lacerda, J.W.; Moura, M.S.; Ali, A.; Vasconcelos, L.G.D.; Sousa Junior, P.T.; Bellete, B.S.; Soares, M.A.; Vieira, L.C.C.; Sampaio, O.M. Metabolomics Analysis of Combretum lanceolatum Roots in the Presence of Its Endophytic Fungi. J. Braz. Chem. Soc. 2023, 34, 234–241. [Google Scholar]
- Wang, Q.; Guo, Q.; Niu, W.; Wu, L.; Gong, W.; Yan, S.; Nishinari, K.; Zhao, M. The pH-responsive phase separation of type-A gelatin and dextran characterized with static multiple light scattering (S-MLS). Food Hydrocoll. 2022, 127, 107503. [Google Scholar] [CrossRef]
- Tang, Y.; Yang, G.; Liu, X.; Qin, L.; Zhai, W.; Fodjo, E.K.; Shen, X.S.; Wang, Y.; Lou, X.Y.; Kong, C. Rapid sample enrichment, novel derivatization, and high sensitivity for determination of 3-chloropropane-1, 2-diol in soy sauce via high-performance liquid chromatography-tandem mass spectrometry. J. Agric. Food Chem. 2023, 71, 15388–15397. [Google Scholar] [CrossRef]
- Kan, Y.; Kan, H.; Bai, Y.; Zhang, S.; Gao, Z. Effective and environmentally safe self-antimildew strategy to simultaneously improve the mildew and water resistances of soybean flour-based adhesives. J. Clean. Prod. 2023, 392, 136319. [Google Scholar] [CrossRef]
- Cui, Y.J.; Liu, P.; Chen, R.Y. Study on the effective constituents of Caulis Spatholobi. China J. Chin. Mater. Med. 2005, 02, 42–44. [Google Scholar]
- Li, Y.J.; Xu, C.T.; Lin, D.D.; Qin, J.K.; Ye, G.J.; Deng, Q.H. Anti-inflammatory polyphenol constituents derived from Cissus pteroclada Hayata. Bioorg. Med. Chem. Lett. 2016, 26, 3425–3428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Su, Y.; Wu, Z.; Gao, X. Bergenin glycosides from Rodgersia aesculifolia. Phytochem. Lett. 2015, 13, 114–118. [Google Scholar] [CrossRef]
- Kopanski, L.; Schnelle, G. Isolation of bergenin from barks of Syzygium cumini. Planta Med. 1988, 54, 572. [Google Scholar] [CrossRef]
- Ye, H.Y.; Chen, C.X.; Hao, X.J. The chemical constituents from Ostryopsis nobilis. Acta Bot. Yunnanica 1996, 18, 473–475. [Google Scholar]
- Frees, D.; Andersen, J.H.; Hemmingsen, L.; Koskenniemi, K.; Bæk, K.T.; Muhammed, M.K.; Gudeta, D.D.; Nyman, T.A.; Sukura, A.; Varmanen, P.; et al. New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J. Proteome Res. 2012, 11, 95–108. [Google Scholar] [CrossRef]
- Gao, P.; Ho, P.L.; Yan, B.; Sze, K.H.; Davies, J.; Kao, R.Y.T. Suppression of Staphylococcus aureus virulence by a small-molecule compound. Proc. Natl. Acad. Sci. USA 2018, 115, 8003–8008. [Google Scholar] [CrossRef]
- Luo, J.; Dong, B.; Wang, K.; Cai, S.; Liu, T.; Cheng, X.; Lei, D.; Chen, Y.; Li, Y.; Kong, J.; et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS ONE 2017, 12, e0176883. [Google Scholar] [CrossRef]
- Cui, L.L.; Zhang, Y.Y.; Shao, W.; Gao, D.M. Analysis of the HPLC fingerprint and QAMS from Pyrrosia species. Ind. Crop. Prod. 2016, 85, 29–37. [Google Scholar] [CrossRef]
- Liu, W.; Liu, Y.; Liu, J.B.; Guo, H.J.; Zheng, L.Z.; Zhou, L.Y.; Zhong, Y.N.; Qin, J.P. Study on the spectrum effect relationship of anti-inflammatory effect of different polar parts of Ampelopsis grossedentata in mice based on grey correlation analysis. Chin. Pharm. 2020, 31, 2382–2386. [Google Scholar]
- Zhang, X.F.; Chen, J.; Yang, J.L.; Shi, Y.P. UPLC-MS/MS analysis for antioxidant components of Lycii Fructus based on spectrum-effect relationship. Talanta 2020, 180, 389–395. [Google Scholar] [CrossRef]
- Zhang, H.; He, X.M.; Wang, X.T.; Yu, B.; Zhao, S.Q.; Jiao, P.L.; Jin, H.W.; Liu, Z.M.; Wang, K.W.; Zhang, L.R.; et al. Design, synthesis and biological activities of piperidinespirooxadiazole derivatives as α7 nicotinic receptor antagonists. Eur. J. Med. Chem. 2020, 207, 112774–112790. [Google Scholar] [CrossRef]















| No. | Strains | Root (100 mg/mL) | Stem (100 mg/mL) | Leaf (100 mg/mL) | Ceftazidime a (1.0 mg/mL) |
|---|---|---|---|---|---|
| 1 | Staphylococcus aureus (ATCC 6538P) | 8.08 ± 0.27 | 6.50 ± 0.33 | 8.98 ± 1.56 | 22.89 ± 2.78 |
| 2 | Bacillus subtilis (ATCC 6633) | 8.60 ± 1.19 | 6.54 ± 0.23 | 7.13 ± 0.72 | 30.07 ± 3.11 |
| 3 | Enterococcus faecalis (ATCC 19433) | 6.15 ± 0.08 | 6.04 ± 0.02 | 6.10 ± 0.07 | 26.42 ± 2.71 |
| 4 | Escherichia coli (CICC 10389) | 6.10 ± 0.06 | 6.41 ± 0.12 | 6.89 ± 0.68 | 29.57 ± 2.63 |
| 5 | Pseudomonas aeruginosa (ATCC 9027) | 8.83 ± 0.11 | 7.13 ± 0.89 | 10.48 ± 1.97 | 28.00 ± 2.74 |
| 6 | Proteus vulgaris (ACCC 11002) | 6.42 ± 0.27 | 6.46 ± 0.32 | 8.71 ± 1.35 | 23.85 ± 2.89 |
| Nystatin b (1.0 mg/mL) | |||||
| 7 | Candida albicans (BNCC 186382) | 10.89 ± 1.58 | 6.11 ± 0.03 | 6.08 ± 0.54 | 25.75 ± 2.67 |
| 8 | Aspergillus flavus (A1142B) | 20.84 ± 1.76 | 7.25 ± 0.62 | 6.15 ± 0.08 | 12.27 ± 1.32 |
| No. | Strains | Root | Stem | Leaf | Ceftazidime a |
|---|---|---|---|---|---|
| 1 | Staphylococcus aureus (ATCC 6538P) | 6.94 ± 0.41 | 8.36 ± 0.29 | 6.77 ± 0.37 | 0.16 ± 0.01 |
| 2 | Bacillus subtilis (ATCC 6633) | 6.87 ± 0.12 | 8.29 ± 0.24 | 7.53 ± 0.33 | 0.04 ± 0.003 |
| 3 | Enterococcus faecalis (ATCC 19433) | 8.33 ± 0.31 | 8.93 ± 0.16 | 8.82 ± 0.18 | 0.08 ± 0.005 |
| 4 | Escherichia coli (CICC 10389) | 8.85 ± 0.14 | 8.41 ± 0.23 | 8.11 ± 0.12 | 0.04 ± 0.003 |
| 5 | Pseudomonas aeruginosa (ATCC 9027) | 6.84 ± 0.15 | 7.55 ± 0.34 | 3.12 ± 0.11 | 0.08 ± 0.006 |
| 6 | Proteus vulgaris (ACCC 11002) | 8.47 ± 0.24 | 8.44 ± 0.32 | 6.94 ± 0.24 | 0.16 ± 0.02 |
| Nystatin b | |||||
| 7 | Candida albicans (BNCC 186382) | 1.56 ± 0.15 | 3.06 ± 0.24 | 3.13 ± 0.12 | 0.05 ± 0.004 |
| 8 | Aspergillus flavus (A1142B) | 0.39 ± 0.023 | 2.77 ± 0.015 | 3.09 ± 0.11 | 0.21 ± 0.02 |
| 8 | Aspergillus flavus (A1142B) | 20.84 ± 1.76 | 7.25 ± 0.62 | 6.15 ± 0.08 | 12.27 ± 1.32 |
| No. | Rt [min] | Name | Molecular Weight | Structures | Formula |
|---|---|---|---|---|---|
| 1 | 19.25 | (−)-gallocatechin | 306.27 |  | C15H14O7 |
| 2 | 23.84 | 11-α-d-galactopyranoside bergenin | 490.41 |  | C20H26O14 |
| 3 | 27.13 | 11-β-d-glucopyranosyl bergenin | 490.41 |  | C20H26O14 |
| 4 | 30.57 | bergenin | 328.27 |  | C14H16O9 |
| 5 | 62.72 | 11-O-galloybergenin | 480.38 |  | C21H20O13 |
| Batch | Root | Stem | Leaf |
|---|---|---|---|
| S1 | 1.000 | 1.000 | 1.000 |
| S2 | 0.998 | 0.997 | 0.997 |
| S3 | 0.999 | 0.994 | 0.580 |
| S4 | 0.998 | 0.620 | 0.674 |
| S5 | 0.988 | 0.997 | 0.975 |
| S6 | 0.979 | 0.996 | 0.986 |
| S7 | 0.983 | 0.996 | 0.979 |
| S8 | 0.999 | 0.995 | 0.920 |
| S9 | 0.998 | 0.996 | 0.995 |
| S10 | 0.998 | 0.543 | 0.959 |
| S11 | 0.998 | 0.543 | 0.959 |
| S12 | 0.998 | 0.694 | 0.751 |
| S13 | 0.999 | 0.528 | 0.959 |
| S14 | 0.987 | 0.890 | 0.700 |
| S15 | 0.997 | 0.904 | 0.600 |
| S16 | 0.992 | 0.643 | 0.775 |
| Reference fingerprint | 0.998 | 0.972 | 0.980 |
| Microorganisms | 11-β-d-Glucopyranosyl-bergenin | 11-α-d-Galacto pyrnside-bergenin | 11-O-Galloybergenin | Bergenin | (−)-Gallocatechin | Ceftazidime a |
|---|---|---|---|---|---|---|
| Staphylococcus aureus (ATCC 6538P) | 9.28 ± 0.93 | 8.13 ± 0.88 | 7.91 ± 0.91 | 6.83 ± 0.74 | 6.54 ± 0.66 | 22.73 ± 2.66 |
| Bacillus subtilis (ATCC 6633) | - | - | - | - | 6.28 ± 0.68 | 30.02 ± 3.32 |
| Enterococcus faecalis (ATCC 19433) | - | - | - | 6.22 ± 0.75 | - | 26.31 ± 2.88 |
| Escherichia coli (CICC 10389) | 6.23 ± 0.72 | 6.08 ± 0.61 | - | - | - | 29.42 ± 2.77 |
| Pseudomonas aeruginosa (ATCC 9027) | 6.19 ± 0.74 | 6.41 ± 0.78 | 7.18 ± 0.87 | 6.11 ± 0.78 | 8.23 ± 0.89 | 28.13 ± 2.81 |
| Proteus vulgaris (ACCC 11002) | - | - | 6.11 ± 0.85 | - | - | 23.52 ± 2.68 |
| Nystatin b | ||||||
| Candida albicans (BNCC 186382) | - | - | 6.34 ± 0.76 | 6.74 ± 0.92 | - | 25.61 ± 2.46 |
| Aspergillus flavus (A1142B) | - | - | 6.17 ± 0.75 | 6.21 ± 0.83 | - | 12.39 ± 1.24 |
| Peak Number | Root | Peak Number | Leaf | ||
|---|---|---|---|---|---|
| Candida albicans | Aspergillus flavus | Pseudomonas aeruginosa | Staphylococcus aureus | ||
| 1 | 0.77 | 0.75 | 1 | 0.83 | 0.82 |
| 4 | 0.79 | 0.78 | 3 | 0.84 | 0.83 |
| 5 | 0.82 | 0.83 | 4 | 0.82 | 0.81 |
| 6 | 0.88 | 0.88 | 5 | 0.83 | 0.85 |
| 9 | 0.82 | 0.82 | 6 | 0.84 | 0.87 |
| 11 | 0.82 | 0.82 | 7 | 0.72 | 0.76 |
| 12 | 0.85 | 0.84 | 8 | 0.77 | 0.75 |
| 15 | 0.83 | 0.83 | 10 | 0.73 | 0.74 |
| 16 | 0.78 | 0.79 | 13 | 0.88 | 0.92 |
| 19 | 0.83 | 0.84 | 16 | 0.86 | 0.85 |
| 21 | 0.81 | 0.79 | 19 | 0.80 | 0.73 |
| 22 | 0.84 | 0.84 | 21 | 0.73 | 0.78 |
| 23 | 0.87 | 0.90 | 22 | 0.86 | 0.84 |
| 25 | 0.73 | 0.71 | 23 | 0.72 | 0.71 |
| 26 | 0.89 | 0.86 | 24 | 0.81 | 0.79 |
| 27 | 0.89 | 0.88 | 26 | 0.75 | 0.73 |
| 28 | 0.68 | 0.70 | |||
| 29 | 0.87 | 0.84 | |||
| No. | Rt (min) | Name | Root | Stem | Leaf | Regression Equation |
|---|---|---|---|---|---|---|
| 1 | 19.25 | (−)-gallocatechin | 5.55 | 3.36 | 2.82 | Y = 13,065x − 294.98 R2 = 0.9995 |
| 2 | 23.84 | 11-α-d-galactopyranoside-bergenin | 0.01 | - | - | Y = 8866x + 92.139 R2 = 0.9994 |
| 3 | 27.13 | 11-β-d-glucopyranosyl-bergenin | 0.21 | 0.38 | 0.16 | Y = 13,851x + 105.93 R2 = 0.9993 |
| 4 | 30.57 | bergenin | 19.11 | 17.80 | 8.10 | Y = 42,625x + 310.13 R2 = 0.9991 |
| 5 | 62.72 | 11-O-galloybergenin | 2.01 | 1.48 | 3.84 | Y = 28,407x − 1084.7 R2 = 0.9990 |
| Compounds | ClpP PRs (3V5e) | LasA PRs (3IT7) | LasB PRs (3DBK) | DNA Gyrase (2XCQ) | DNA Ligase (3JSN) | MurF Ligase (4CVL) |
|---|---|---|---|---|---|---|
| 11-β-d-glucopyranosyl-bergenin | −9.84 | −1.87 | −3.58 | −1.18 | −1.82 | −1.53 |
| 11-α-d-galactopyranoside-bergenin | −8.58 | −2.93 | −3.90 | −1.56 | −2.28 | −4.19 |
| 11-O-galloybergenin | −8.34 | −5.08 | −5.66 | −3.41 | −4.91 | −4.73 |
| bergenin | −7.79 | −5.49 | −5.59 | −4.17 | −4.22 | −1.21 |
| (−)-gallocatechin | −7.65 | −6.61 | −6.81 | −4.55 | −1.14 | −5.03 |
| Ceftazidime a | −3.98 | −3.17 | −3.86 | −5.63 | −6.72 | −3.19 |
| Compounds | SQS PRs (7WG1) | 1,3-β-Glucan Synthase (8JZN) | Chitin Synthase (7STL) |
|---|---|---|---|
| 11-β-d-glucopyranosyl-bergenin | −1.39 | −2.23 | −4.51 |
| 11-α-d-galactopyranoside-bergenin | −1.52 | −2.42 | −4.13 |
| 11-O-galloybergenin | −4.23 | −1.18 | −2.92 |
| bergenin | −4.61 | −4.93 | −2.56 |
| (−)-gallocatechin | −3.02 | −4.62 | −1.33 |
| Nystatin a | −3.19 | −6.38 | −3.11 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, C.; Wang, C.; Zhou, Y.; Hu, T.; Zhang, Y.; Lv, X.; Li, J.; Zhou, Y. Discovery of Potential Anti-Microbial Molecules and Spectrum Correlation Effect of Ardisia crenata Sims via High-Performance Liquid Chromatography Fingerprints and Molecular Docking. Molecules 2024, 29, 1178. https://doi.org/10.3390/molecules29051178
Zhao C, Wang C, Zhou Y, Hu T, Zhang Y, Lv X, Li J, Zhou Y. Discovery of Potential Anti-Microbial Molecules and Spectrum Correlation Effect of Ardisia crenata Sims via High-Performance Liquid Chromatography Fingerprints and Molecular Docking. Molecules. 2024; 29(5):1178. https://doi.org/10.3390/molecules29051178
Chicago/Turabian StyleZhao, Chunli, Changbin Wang, Yongqiang Zhou, Tao Hu, Yan Zhang, Xiang Lv, Jiaxin Li, and Ying Zhou. 2024. "Discovery of Potential Anti-Microbial Molecules and Spectrum Correlation Effect of Ardisia crenata Sims via High-Performance Liquid Chromatography Fingerprints and Molecular Docking" Molecules 29, no. 5: 1178. https://doi.org/10.3390/molecules29051178
APA StyleZhao, C., Wang, C., Zhou, Y., Hu, T., Zhang, Y., Lv, X., Li, J., & Zhou, Y. (2024). Discovery of Potential Anti-Microbial Molecules and Spectrum Correlation Effect of Ardisia crenata Sims via High-Performance Liquid Chromatography Fingerprints and Molecular Docking. Molecules, 29(5), 1178. https://doi.org/10.3390/molecules29051178