Association Study between BDNF Gene Polymorphisms and Autism by Three-Dimensional Gel-Based Microarray
Abstract
:1. Introduction
2. Results and Discussion
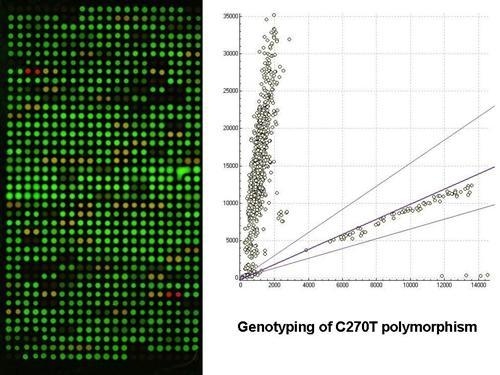
2.1. Gel-based microarray for genotyping
2.2. Association study
3. Experimental Section
3.1. Subjects
3.2. Genotyping
3.2.1. PCR Amplification
3.2.2. Immobilization of PCR products
3.2.3. Hybridization
3.2.4. Post-hybridization
3.2.5. Scanning and genotyping
3.3. Statistical analysis
4. Conclusions
Acknowledgments
References and Notes
- Hoggart, CJ; Clark, TG; De Lorio, M; Whittaker, JC; Balding, DJ. Genome-wide significance for dense SNP and resequencing data. Genet. Epidemiol 2008, 32, 179–185. [Google Scholar]
- Hinds, DA; Stuve, LL; Nilsen, GB; Halperin, E; Eskin, E; Ballinger, DG; Frazer, KA; Cox, DR. Whole-genome patterns of common DNA variation in three human populations. Science 2005, 307, 1072–1079. [Google Scholar]
- Saxena, R; Voight, BF; Lyssenko, V; Burtt, NP; de Bakker, PIW; Chen, H; Roix, JJ; Kathiresan, S; Hirschhorn, JN; Daly, MJ; Hughes, TE; Groop, L; Altshuler, D; Almgren, P; Florez, JC; Meyer, J; Ardlie, K; Bostrom, KB; Isomaa, B; Lettre, G; Lindblad, U; Lyon, HN; Melander, O; Newton-Cheh, C; Nilsson, P; Orho-Melander, M; Rastam, L; Speliotes, EK; Taskinen, MR; Tuomi, T; Guiducci, C; Berglund, A; Carlson, J; Gianniny, L; Hackett, R; Hall, L; Holmkvist, J; Laurila, E; Sjogren, M; Sterner, M; Surti, A; Svensson, M; Svensson, M; Tewhey, R; Blumenstiel, B; Parkin, M; DeFelice, M; Barry, R; Brodeur, W; Camarata, J; Chia, N; Fava, M; Gibbons, J; Handsaker, B; Healy, C; Nguyen, K; Gates, C; Sougnez, C; Gage, D; Nizzari, M; Gabriel, SB; Chirn, GW; Ma, QC; Parikh, H; Richardson, D; Ricke, D; Purcell, S; In, DGIB; Res, NIB. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007, 316, 1331–1336. [Google Scholar]
- Sham, PC; Cherny, SS; Kao, PYP; Song, YQ; Chan, D; Cheung, KMC. Whole-genome association studies of complex diseases. Curr. Orthopaed 2008, 22, 251–258. [Google Scholar]
- Li, S; Liu, HN; Wang, ZF; Ji, MJ; Guo, YF; He, NY; Dai, YB. A novel in situ magnetic particle PCR based high-throughput genotyping method using universal tags. Prog. Biochem. Biophys 2007, 34, 1107–1112. [Google Scholar]
- Liu, HN; Li, S; Ji, MJ; Nie, LB; Chen, JR; Miao, YQ; He, NY. Fabrication and application of single nucleotide polymorphisms library on magnetic nanoparticles using adaptor PCR. J. Nanosci. Nanotechnol 2008, 8, 405–409. [Google Scholar]
- Van Tassell, CP; Smith, TPL; Matukumalli, LK; Taylor, JF; Schnabel, RD; Lawley, CT; Haudenschild, CD; Moore, SS; Warren, WC; Sonstegard, TS. SNP discovery and allele frequency estimation by deep sequencing of reduced representation libraries. Nat. Methods 2008, 5, 247–252. [Google Scholar]
- Dumaual, C; Miao, X; Daly, TM; Bruckner, C; Njau, R; Fu, DJ; Close-Kirkwood, S; Bauer, N; Watanabe, N; Hardenbol, P; Hockett, RD. Comprehensive assessment of metabolic enzyme and transporter genes using the Affymetrix (R) Targeted Genotyping System. Pharmacogenomics 2007, 8, 293–305. [Google Scholar]
- Kaller, M; Lundeberg, J; Ahmadian, A. Arrayed identification of DNA signatures. Expert Rev. Mol. Diagn 2007, 7, 65–76. [Google Scholar]
- Ragoussis, J; Elvidge, G. Affymetrix GeneChip (R) system: Moving from research to the clinic. Expert Rev. Mol. Diagn 2006, 6, 145–152. [Google Scholar]
- Ji, MJ; Hou, P; Li, S; He, NY; Lu, ZH. Microarray-based method for genotyping of functional single nucleotide polymorphisms using dual-color fluorescence hybridization. Mutat. Res-Fundam Mol. Mech. Mut 2004, 548, 97–105. [Google Scholar]
- Xiao, PF; Cheng, L; Wan, Y; Sun, BL; Chen, ZZ; Zhang, SY; Zhang, CZ; Zhou, GH; Lu, ZH. An improved gel-based DNA microarray method for detecting single nucleotide mismatch. Electrophoresis 2006, 27, 3904–3915. [Google Scholar]
- Xiao, P; Huang, H; Zhou, G; Lu, Z. Gel immobilization of acrylamide-modified single-stranded DNA template for pyrosequencing. Electrophoresis 2007, 28, 1903–1912. [Google Scholar]
- Kim, SJ; Brune, CW; Kistner, EO; Christian, SL; Courchesne, EH; Cox, NJ; Cook, EH. Transmission Disequilibrium, Testing of the Chromosome 15q11-q13 Region in Autism. Am. J. Med. Genet. Part B 2008, 147B, 1116–1125. [Google Scholar]
- Ke, XY; Cheng, L; Zou, B; Zhou, Y; Chen, P; Sun, BL; Hang, YY; Zhang, MH; Sun, Y; Zheng, YZ; Wang, MJ; Lu, ZH. Study on association between autism and genotypes of tryptophan 2,3-dioxygenase in Chinese Han population. Prog. Post-Genome Technol 2007, 494, 441–443. [Google Scholar]
- Muhle, R; Trentacoste, S; Rapin, VI. The genetics of autism. Pediatrics 2004, 113, e472–e486. [Google Scholar]
- Kozisek, ME; Middlemas, D; Bylund, DB. Brain-derived neurotrophic factor and its receptor tropomyosin-related kinase B in the mechanism of action of antidepressant therapies. Pharmacol. Ther 2008, 117, 30–51. [Google Scholar]
- Schumacher, J; Jamra, RA; Becker, T; Ohlraun, S; Klopp, N; Binder, EB; Schulze, TG; Deschner, M; Schmal, C; Hofels, S; Zobel, A; Illig, T; Propping, P; Holsboer, F; Rietschel, M; Nothen, MM; Cichon, S. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol. Psychiat 2005, 58, 307–314. [Google Scholar]
- Nishimura, K; Nakamura, K; Anitha, A; Yamada, K; Tsujii, M; Iwayama, Y; Hattori, E; Toyota, T; Takei, N; Miyachi, T; Iwata, Y; Suzuki, K; Matsuzaki, H; Kawai, M; Sekine, Y; Tsuchiya, K; Sugihara, G; Suda, S; Ouchi, Y; Sugiyama, T; Yoshikawa, T; Mori, N. Genetic analyses of the brain-derived neurotrophic factor (BDNF) gene in autism. Biochem. Biophys. Res. Commun 2007, 356, 200–206. [Google Scholar]
- Rehman, FN; Audeh, M; Abrams, ES; Hammond, PW; Kenney, M; Boles, TC. Immobilization of acrylamide-modified oligonucleotides by co-polymerization. Nucleic Acids Res 1999, 27, 649–655. [Google Scholar]
- Thoenen, H. Neurotrophins and neuronal plasticity. Science 1995, 270, 593–598. [Google Scholar]
- Schinder, AF; Poo, M. The neurotrophin hypothesis for synaptic plasticity. Trends Neurosci 2001, 23, 639–645. [Google Scholar]
- Perry, EK; Lee, MLW; Martin-Ruiz, CM; Court, JA; Volsen, SG; Merrit, J; Folly, E; Iversen, PE; Bauman, ML; Perry, RH. Cholinergic activity in autism: abnormalities in the cerebral cortex and basal forebrain. Am. J. Psychiat 2001, 158, 1058–1066. [Google Scholar]
- Nelson, KB; Grether, JK; Croen, LA; Dambrosia, JM; Dickens, BF; Jelliffe, LL; Hansen, RL; Phillips, TM. Neuropeptides and neurotrophins in neonatal blood of children with autism or mental retardation. Ann. Neurol 2001, 49, 597–606. [Google Scholar]
- Connolly, AM; Chez, M; Streif, EM; Keeling, RM; Golumbek, PT; Kwon, JM; Riviello, JJ; Robinson, RG; Neuman, RJ; Deuel, RMK. Brain-derived neurotrophic factor and autoantibodies to neural antigens in sera of children with autistic spectrum disorders, Landau-Kleffner syndrome, and epilepsy. Biol. Psychiat 2006, 59, 354–363. [Google Scholar]
- Miyazaki, K; Narita, N; Sakuta, R; Miyahara, T; Naruse, H; Okado, N; Narita, M. Serum neurotrophin concentrations in autism and mental retardation: a pilot study. Brain Dev 2004, 26, 292–295. [Google Scholar]
- Chugani, DC; Muzik, O; Behen, M; Rothermel, R; Janisse, JJ; Lee, J; Chugani, HT. Developmental changes in brain serotonin synthesis capacity in autistic and nonautistic children. Ann. Neurol 1999, 45, 287–295. [Google Scholar]
- Chugani, DC; Muzik, O; Rothermel, R; Behen, M; Chakraborty, P; Mangner, T; Da Silva, EA; Chugani, HT. Altered serotonin synthesis in the dentatothalamocortical pathway in autistic boys. Ann. Neurol 1997, 42, 666–669. [Google Scholar]
- Matrisciano, F; Bonaccorso, S; Ricciardi, A; Scaccianoce, S; Panaccione, I; Wang, L; Ruberto, A; Tatarelli, R; Nicoletti, F; Girardi, P; Shelton, RC. Changes in BDNF serum levels in patients with major depression disorder (MDD) after 6 months treatment with sertraline, escitalopram, or venlafaxine. J. Psychiat. Res 2008, 43, 247–254. [Google Scholar]
- Hall, D; Dhilla, A; Charalambous, A; Gogos, JA; Karayiorgou, M. Sequence variants of the brain-derived neurotrophic factor (BDNF) gene are strongly associated with obsessive-compulsive disorder. Am. J. Hum. Genet 2003, 73, 370–376. [Google Scholar]
- Kent, L; Green, E; Hawi, Z; Kirley, A; Dudbridge, F; Lowe, N; Raybould, R; Langley, K; Bray, N; Fitzgerald, M. Association of the paternally transmitted copy of common Valine allele of the Val66Met polymorphism of the brain-derived neurotrophic factor (BDNF) gene with susceptibility to ADHD. Mol. Psychiat 2005, 10, 939–943. [Google Scholar]
- Xu, X; Mill, J; Zhou, K; Brookes, K; Chen, CK; Asherson, P. Family-based association study between brain-derived neurotrophic factor gene polymorphisms and attention deficit hyperactivity disorder in UK and Taiwanese samples. Am. J. Med. Genet. B 2007, 144B, 83–86. [Google Scholar]
- Lang, UE; Hellweg, R; Kalus, P; Bajbouj, M; Lenzen, KP; Sander, T; Kunz, D; Gallinat, J. Association of a functional BDNF polymorphism and anxiety-related personality traits. Psychopharmacology (Berlin) 2005, 180, 95–99. [Google Scholar]
- Maisonpierre, PC; Le Beau, MM; Espinosa, R, 3rd; Ip, NY; Belluscio, L; De La Monte, SM; Squinto, S; Furth, ME; Yancopoulos, GD. Human and rat brain-derived neurotrophic factor and neurotrophin-3: Gene structures, distributions, and chromosomal localizations. Genomics 1991, 10, 558–568. [Google Scholar]
- Szekeres, G; Juhasz, A; Rimanoczy, A; Keri, S; Janka, Z. The C270T polymorphism of the brain-derived neurotrophic factor gene is associated with schizophrenia. Schizophr Res 2003, 65, 15–18. [Google Scholar]
- Shimizu, E; Hashimoto, K; Iyo, M. Ethnic difference of the BDNF 196G/A (val66met) polymorphism frequencies: The possibility to explain ethnic mental traits. Am. J. Med. Genet. B 2004, 126B, 122–123. [Google Scholar]
- Kaufman, J; Yang, BZ; Douglas-Palumberi, H; Grasso, D; Lipschitz, D; Houshyar, S; Krystal, JH; Gelernter, J. Brain-derived neurotrophic factor–5-HTTLPR gene interactions and environmental modifiers of depression in children. Biol. Psychiatry 2006, 59, 673–680. [Google Scholar]
- Egan, MF; Kojima, M; Callicott, JH; Goldberg, TE; Kolachana, BS; Bertolino, A; Zaitsev, E; Gold, B; Goldman, D; Dean, M; Lu, B; Weinberger, DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003, 112, 257–269. [Google Scholar]
- Hariri, AR; Goldberg, TE; Mattay, VS; Kolachana, BS; Callicott, JH; Egan, MF; Weinberger, DR. Brain-derived neurotrophic factor val66met polymorphism affects human memory-related hippocampal activity and predicts memory performance. J. Neurosci 2003, 23, 6690–6694. [Google Scholar]
- Yong, Y; Lin, HE. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 2005, 15, 97–98. [Google Scholar]



| SNP | Group | HWE | Genotype (frequency %) | p value | Allele (frequency %) | p value | ORa | 95%CIb | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | C | G | |||||||
| rs988748 | Autism | 0.175 | 33(0.266) | 69(0.556) | 22(0.177) | 0.228 | 135(0.544) | 113(0.456) | 0.807 | 1.045 | 0.732~1.492 |
| Control | 0.293 | 37(0.308) | 54(0.450) | 29(0.242) | 128(0.533) | 112(0.467) | |||||
| AA | AT | TT | A | T | |||||||
| rs2049046 | Autism | 0.208 | 28(0.226) | 69(0.556) | 27(0.218) | 0.191 | 125(0.504) | 123(0.496) | 0.710 | 0.935 | 0.655~1.334 |
| Control | 0.207 | 36(0.300) | 53(0.442) | 31(0.258) | 125(0.521) | 115(0.479) | |||||
| CC | CT | TT | C | T | |||||||
| C270T | Autism | 0.876 | 96(0.774) | 26(0.210) | 2(0.016) | 0.020 | 218(0.879) | 30(0.121) | 0.005 | 0.382 | 0.191~0.766 |
| Control | 0.564 | 108(0.900) | 12(0.100) | 0(0.000) | 228(0.950) | 12(0.050) | |||||
| AA | AG | GG | A | G | |||||||
| rs6265 | Autism | 0.129 | 22(0.177) | 70(0.565) | 32(0.258) | 0.274 | 114(0.460) | 134(0.540) | 0.734 | 0.940 | 0.658~1.342 |
| Control | 0.481 | 29(0.242) | 56(0.467) | 35(0.292) | 114(0.475) | 126(0.525) | |||||
| Haplotype | case | control | chi | p value | OR[95%CI] |
|---|---|---|---|---|---|
| G A C A | 96.09(0.387) | 107.30(0.447) | 1.300 | 0.2543281 | 0.808 [0.560~1.166] |
| C A C G | 9.21(0.037) | 13.51(0.056) | 0.901 | 0.342672 | 0.696 [0.293~1.650] |
| C T C G | 88.96(0.359) | 104.14(0.434) | 2.296 | 0.129761 | 0.752 [0.520~1.088] |
| C T T G | 21.99(0.089) | 7.81(0.033) | 7.056 | 0.007923 | 2.972 [1.286~6.868] |
| C A C A | 10.26(0.041) | 0.00(0.000) | 10.382 | 0.001281 | — |
| G T C G | 8.58(0.035) | 0.00(0.000) | 8.767 | 0.003082 | |
| Global result: Total control = 240.0, total case = 248.0, χ2 = 28.194, df = 5, p = 3.44e-005 | |||||
| SNP ID | Primer sequence (5′-3′) | Fragment size (bp) | Probe sequence |
|---|---|---|---|
| rs988748 | F: 5′-TAGGGTTCCTCCAGTCCTTT | 250 | 5′-Cy3-GGGTCTCTGGGGT |
| R: ’-Acryl-CAGCACAGATGGCAGAGTTTA | 5′-Cy5-GGGTCTGTGGGGT | ||
| rs2049046 | F: 5′-CAGGAGGAGGGACCTTCATT | 293 | 5′-Cy3-CCAGGGACTCCAA |
| R: 5′-Acryl-AGCCTTTCGGGTTCTCATTT | 5′-Cy5-CCAGGGTCTCCAA | ||
| C270T | F: 5′-CAGAGGAGCCAGCCCGGTGCG | 213 | 5′-Cy3-CTCCACCTCCTGC |
| R: 5′-Acryl-CTCCTGCACCAAGCCCCATTC | 5′-Cy5-CTCCACTTCCTGC | ||
| rs6265 | F: 5′-AAACATCCGAGGACAAGGTG | 246 | 5′-Cy3-GAACACATGATAG |
| R: 5′-Acryl-AGAAGAGGAGGCTCCAAAGG | 5′-Cy5-GAACACGTGATAG |
© 2009 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Cheng, L.; Ge, Q.; Xiao, P.; Sun, B.; Ke, X.; Bai, Y.; Lu, Z. Association Study between BDNF Gene Polymorphisms and Autism by Three-Dimensional Gel-Based Microarray. Int. J. Mol. Sci. 2009, 10, 2487-2500. https://doi.org/10.3390/ijms10062487
Cheng L, Ge Q, Xiao P, Sun B, Ke X, Bai Y, Lu Z. Association Study between BDNF Gene Polymorphisms and Autism by Three-Dimensional Gel-Based Microarray. International Journal of Molecular Sciences. 2009; 10(6):2487-2500. https://doi.org/10.3390/ijms10062487
Chicago/Turabian StyleCheng, Lu, Qinyu Ge, Pengfeng Xiao, Beili Sun, Xiaoyan Ke, Yunfei Bai, and Zuhong Lu. 2009. "Association Study between BDNF Gene Polymorphisms and Autism by Three-Dimensional Gel-Based Microarray" International Journal of Molecular Sciences 10, no. 6: 2487-2500. https://doi.org/10.3390/ijms10062487
APA StyleCheng, L., Ge, Q., Xiao, P., Sun, B., Ke, X., Bai, Y., & Lu, Z. (2009). Association Study between BDNF Gene Polymorphisms and Autism by Three-Dimensional Gel-Based Microarray. International Journal of Molecular Sciences, 10(6), 2487-2500. https://doi.org/10.3390/ijms10062487