1. Introduction
Computing is the study of natural and artificial information processes [
1]. Natural computing includes the implementation of computational paradigms abstracted from natural phenomena either on traditional electronic hardware or on alternative physical media such as in biomolecular (DNA, RNA) computing [
2]. Biocompatible systems that can process information are sought for future biomedical applications. Biomolecular systems are potentially important for these and more general computational applications because of their intrinsically high information storage capabilities and parallelism.
DNA is used to build synthetic molecular machines because the simplicity of its structure and interactions allows control of its assembly through information stored in nucleotide sequences [
3,
4]. The Watson–Crick double helix is formed by linking together two antiparallel strands of DNA that have complementary base sequences, a process known as hybridization. Attractive interactions between complementary nucleotides contribute to the stability of the structure: adenine (A) on one strand pairs with thymine (T) on the other, while cystosine (C) pairs with guanine (G). It is the remarkable specificity of the interactions between these complementary nucleotides, together with the availability of routine commercial synthesis, that allows DNA to be used as engineering material with which to design complex systems and structures capable of self-assembly and parallel operation [
3,
4]. This versatile molecule has been used to design machines [
5–
13], finite automata [
14,
15], logic gates [
16,
17], reaction networks [
18–
20] and logic circuits [
21,
22], amongst many other structures and dynamic systems. Refer to [
3,
4,
23] for reviews of DNA devices and machines.
Here we present preliminary work towards an autonomous, synthetic DNA network, comprised solely of DNA molecules. This DNA network implements computing paradigms abstracted from natural cellular biochemical reaction networks.
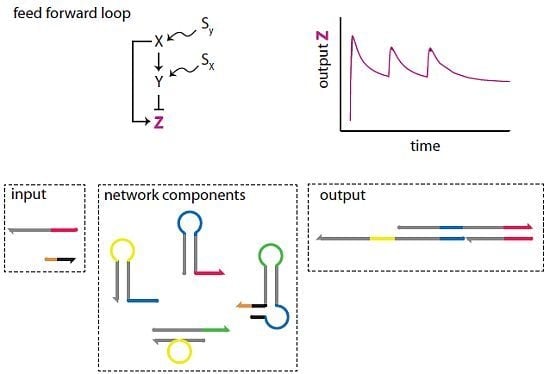
Cellular regulation is achieved through complex networks of interactions among biochemicals and cellular structures. Recurrent network motifs, classifiable in terms of function, architecture, dynamics, or biochemical process [
24–
26], have been identified. Alon and colleagues investigated transcription networks in the bacterium
E. coli and the yeast
S. cerevisiae whose information processing role is to determine the rate of production of specific proteins as a function of the environment [
25,
26]. In these networks, the nodes are genes and the edges represent transcriptional regulation of one gene by the protein product of another gene. Two important motifs in
E. coli and
S. cerevisiae transcription networks are the type-1 coherent feed-forward loop (C1-FFL) and the type-1 incoherent feed-forward loop (I1-FFL). In the C1-FFL (
Figure 1a) both paths are positive: X activates both Z and an activator of Z. In the I1-FFL (
Figure 1b), the direct path is positive and the indirect path is negative,
i.e., X activates Z and an inhibitor of Z. Experimental and computational approaches have shown that the C1-FFL shows sign-sensitive delay that can protect against brief input fluctuations [
27], whereas the I1-FFL is responsible for functions such as pulse generation [
28], adaptation [
29,
30], fold-change detection [
31] and amplitude filtering [
32].
Figure 1c shows an example of pulse generation using the I1-FFL network motif.
Other networks have been investigated and display interesting information processing properties. Acar
et al. [
34] investigate inducibility and network-dosage invariance (
i.e., invariance to the number of copies of a gene network in a cell) in the yeast galactose network. Their results revealed that, in general, the presence of two network components, one positive and one negative regulator, is the minimal requirement for network-dosage invariance.
In this work, we develop an implementation of an information processing paradigm abstracted from the yeast galactose network using DNA molecular computation. In particular, we aim to show that a DNA system can be designed and programmed to implement an information processing function that is robust to changes in network dosage and is thus capable of contributing to network behaviour that is reliable despite the stochasticity inherent in molecular systems. We choose to implement, on the DNA network, the basic information processing function of the I1-FFL: pulse generation. We note that the I1-FFL network motif [
27] has two components, one positive and one negative regulator, and thus satisfies the minimal requirement for a network whose activity is robust to changes in network dosage [
34].
The paper is organized as follows. Section 2 discusses the toolbox, i.e., the DNA processes and structures from which our DNA network is composed. The network itself is described in Section 3. Section 4 contains the results of numerical simulations, and conclusions are presented in Section 5.
3. The DNA Network Design
The DNA network is shown in
Figure 4. It is abstracted from the topology and functionality of the I1-FFL network motif [
27] and network-dosage invariance of the galactose signalling pathway of yeast [
34]. Its abstract representation is identical to the I1-FFL shown in
Figure 1b. In our DNA implementation, the nodes are complexes of DNA strands. The network has two inputs,
Ix and
Ky, and its activity is measured by the concentration of the output
Z*. Its design is based on the principle of toehold-mediated strand displacement (
Figure 2) and the gated hairpin structure (
Figure 3).
To initiate both positive and negative reaction pathways, input
Ix reacts with hairpin
Hx (as described in
Figure 3a) forming
X* and allowing access to the toehold sequestered in the loop of
Hx (blue). Following the positive reaction pathway,
X* can then react with hairpin
Hz to form
Z*, in which the toehold sequestered in the loop of
Hz (yellow) is activated.
Independently, and in parallel, the second input, strand
Ky, activates the gated hairpin loop
Gy (as described in
Figure 3b), forming
Gy*. The latter reacts with
X* in the negative regulatory pathway, opening the hairpin and forming product
Y*.
Y* binds to double-stranded complex
Aux in a reaction mediated by its exposed toehold (green), displacing strand
Inh from
Aux and forming
Waste. The displaced single strand
Inh hybridizes to
Z*, forming the structure
InhZ in which the domain that encodes the active state of
Z* is inhibited.
The output of the network is measured by the concentration of Z*. The toehold activated by production of Z* (the yellow loop domain that is sequestered in hairpin Hz) is reactive and could be used to cascade downstream reactions. It could act as the input signal to another network or could be designed to regulate the production of its own initiator, input strand Ix, providing feedback to the network.
The gated hairpin loop
Gy is not active unless input strand
Ky is present to open its gate (
Figure 3c). In the absence of input
Ky, the network only has the positive reaction pathway and input
Ix reacts stoichiometrically to produce output
Z*. In the presence of
Ky, through production of
Gy*, a proportion of
Ix is diverted to the negative reaction pathway, leading to inhibition of
Z*. The use of the gated hairpin therefore adds an independent control to the output of the network.
Because the two reaction pathways run in parallel, it is important to consider the effects of the timing of the inputs on the network behaviour. We consider two scenarios. In Scenario A, input Ky is added first, such that its reaction with Gy to form Gy* is substantially complete before the second input Ix is added. In Scenario B, both inputs are added simultaneously. Two effects can contribute to a transient overproduction of output Z* that is later compensated by production of Inh, resulting in a pulse of Z*. In Scenario B, production of intermediate Gy* does not begin until both inputs are added, so the initial reaction rate of intermediate X* with Gy*, corresponding to entry into the negative pathway, is zero: this guarantees a pulse of Z*. Even in Scenario A, in which both pathways are active when Ix is added, a relative delay in the production of Inh resulting from the greater length of the negative pathway gives rise to pulse production.
This network motif is designed to operate far-from-equilibrium, as are the natural biochemical networks that inspired it. The ratios between forward and reverse reaction rates are determined by the free energy changes in each reaction and can be made large by design. Slow reverse reactions will have little effect on transient phenomena (pulse generation). However, if a sufficiently long time is allowed to elapse then reverse reactions—however slow—will ensure that the final state of the system is in thermodynamic equilibrium, independent of the details of reaction pathways. If the output of the network motif cascades forward to actuate downstream processes that are similarly far-from-equilibrium then this equilibrium state is never relevant to its operation.
3.1. Programming the DNA Network
In order to achieve a steady-state concentration of output
Z* that is invariant to network dosage,
i.e., to the concentrations of network components, it is necessary to ensure that the ratio between the time-integrated quantities of input
Ix (and therefore of intermediate
X*) that flow through the positive and negative reaction pathways is invariant. We assume below that the concentrations of network components
Hx,
Hz,
Gy and
Aux and of input
Ky are in sufficient excess over the initial concentration of input
Ix that perturbations resulting from the reactions triggered by addition of
Ix are small. The rate of activation of the negative pathway depends on the concentration of intermediate
Gy* which is, in general, time-dependent and a function of the input
Ky. In Scenario A, when all
Gy* is formed before
Ix is added, the time-dependence of the production of
Ky plays no part in the behaviour of the network. Network-dosage invariance of the steady-state output is achieved if the concentrations of
Hz and the smaller of the concentrations of
Gy and of
Ky are scaled together ensuring a constant branching ratio between the two reaction pathways. In Scenario B,
Ky and
Ix are added simultaneously and the time-dependence of the production of
Gy* does affect the output of the network. In this case, network-dosage invariance is achieved if the concentrations of
Hx,
Hz,
Gy and
Ky are scaled together (see
Supplementary Material). Note that the dynamic component of the network output (the pulse) is not, in general, network-dosage invariant.
The network can be configured in three ways—positive, balanced and negative—defined by its behaviour in Scenario A. The branching ratio between positive and negative pathways is determined by the concentrations of Gy* and Hz which compete for reaction with intermediate X*. In a balanced network, the time-integrated branching ratio is 1:1 and addition of input Ix has no effect on the steady-state concentration of output Z*. In an unbalanced network the proportions of Ix that enter the positive and negative pathways are unequal with the result that the output concentration of Z* is changed by addition of Ix: it can be increased or decreased (though not, of course, below zero), depending on the relative concentrations of Gy* and Hz.
4. Simulation Results
We have investigated the behaviour of the proposed DNA network through chemical kinetics simulations. The network was modelled by the following ordinary differential equations:
All reactions, with the exception of the reaction of
Inh with
Z*, involve toehold-mediated strand displacement to open a secondary structure loop. Rate constants
k1 to
k6 are set at 10
5M−1s−1 [
35,
38]. We have assumed that all reactions are irreversible (rates of reverse reactions can be six orders of magnitude slower for appropriate toehold lengths [
40]). Initial concentrations of DNA molecules are specified in
Supplementary Material, Table 1: concentrations of network components are of the order of 1
μM. Simulations were performed in Matlab [
41] using the ODE solver ode15s.
Simulation results are shown in
Figure 5. The graphs show the concentration of the network output
Z* as a function of time. Unless stated otherwise, the initial concentration of input
Ix is set to 100 nM. All network components are present at t = 0 s.
Figure 5a shows simulation results for Scenario A (Section 3): input
Ky is added first, at t = 0 s, and input
Ix is added at t = 700 s when the reaction of
Ky with
Gy to produce
Gy* is substantially complete. Simulation S1 is unbalanced positively,
i.e., the initial concentration of
Hz is greater than that of
Gy resulting in a non-zero steady-state concentration of output
Z*. In simulation 2S1, the initial concentrations of all network components are doubled: the concentration of
Ky is greater than that of
Gy in both cases. The two networks generate similar pulses, and the steady-state outputs of the two networks are the same, demonstrating that the steady-state output of the DNA network is robust to changes in network dosage. The same network behaviour—pulse generation and network-dosage invariant steady-state output—is displayed for balanced (S2 and 2S2) and negative (S3 and 2S3) networks.
Figure 5b shows simulation results for Scenario B: inputs
Ix and
Ky are delivered simultaneously at t = 0 s. S4-S6 correspond to positive, balanced and negative networks and 2S4–2S6 to the same networks in which the concentrations of network components and input
Ky are doubled. Again, under these conditions the steady-state component of the output is robust to the change in network dosage. Note that, as expected, the steady-state output of the balanced network (S5) is different from the zero output of the same network in Scenario A (S2,
Figure 5a): this asymmetry is a result of the initial unbalance in the network during production of
Gy*. Also shown in
Figure 5b is the result of simulation S7 in which all initial concentrations are the same as in 2S5 with the exception of the concentration of
Ky, which is as in S5 (half that in 2S5). The outputs of 2S5 and S7 are different, as expected, demonstrating that the concentration of input
Ky must be scaled with those of network components in order to achieve network-dosage invariance.
The balanced network has another robust behaviour. Once input
Ky has had time to react—whether in Scenario A or Scenario B—the steady-state level of output
Z* is unchanged by subsequent addition of small quantities of input
Ix. Once a steady (and balanced) concentration of
Gy* is established, these subsequent stimuli
Ix cause equal activation of the positive and negative reaction pathways, resulting in a pulse of
Z* but no change in its steady-state concentration.
Figure 5c shows the effects of subsequent additions of input
Ix. Simulation S8 is the same as Simulation S5, except that two further quantities of
Ix were added after the initial pulse of output
Z* had died away. (For a plot of the state space see
Figure 2 in
Supplementary Material.) The second and third additions of
Ix have no effect on the steady-state output of the network, as expected. The results of Simulation S9 are also shown: S9 has the same quantity of input
Ix added as in Simulation S8, except that it is all added at once at t = 0 s (initial conditions are otherwise the same as Simulations 5 and 8). The steady-state output in Simulation S9 is different from S5 and S8, demonstrating that the lack of effect of the later additions of
Ix is a result of the timing of the inputs and not of saturation of the output of the network.
Figures 3–
5 in
Supplementary Material present additional simulation results showing robustness of output to subsequent stimuli, the limits of the desired network behaviour and results of extreme imbalance between the activation and repression pathways.