From Evolution to Pathogenesis: The Link Between β-Barrel Assembly Machineries in the Outer Membrane of Mitochondria and Gram-Negative Bacteria
Abstract
:1. Basic Features of the Bacterial and Mitochondrial Membranes
2. β-Barrel Proteins and the Assembly in the Bacterial and Mitochondrial Outer Membrane
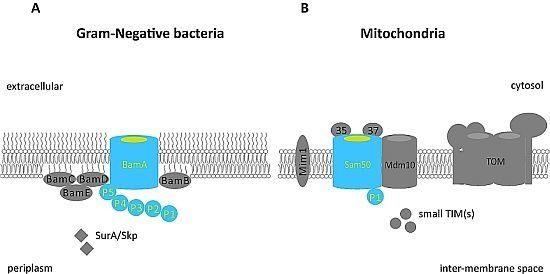
3. The β-Barrel Assembly Machinery in Gram-Negative Bacteria
4. The Sorting and Assembly Machinery (SAM) in Mitochondria
5. How Functionally Conserved Are the BAM and SAM?
6. β-Barrels—Evolution and Pathogenesis
7. Concluding Remarks and Future Directions
References
- Koebnik, R.; Locher, K.P.; van Gelder, P. Structure and function of bacterial outer membrane proteins: Barrels in a nutshell. Mol. Microbiol 2000, 37, 239–253. [Google Scholar]
- Bos, M.P.; Robert, V.; Tommassen, J. Biogenesis of the gram-negative bacterial outer membrane. Annu. Rev. Microbiol 2007, 61, 191–214. [Google Scholar]
- Gray, M.W.; Burger, G.; Lang, B.F. Mitochondrial evolution. Science 1999, 283, 1476–1481. [Google Scholar]
- Yang, D.; Oyaizu, Y.; Oyaizu, H.; Olsen, G.J.; Woese, C.R. Mitochondrial origins. Proc. Natl. Acad. Sci. USA 1985, 82, 4443–4447. [Google Scholar]
- Gray, M.W. The incredible shrinking organelle. EMBO Rep 2011, 12. [Google Scholar] [CrossRef]
- Endo, T.; Yamano, K. Transport of proteins across or into the mitochondrial outer membrane. Biochim. Biophys. Acta 2010, 1803, 706–714. [Google Scholar]
- Fairman, J.W.; Noinaj, N.; Buchanan, S.K. The structural biology of β-barrel membrane proteins: A summary of recent reports. Curr. Opin. Struct. Biol 2011, 21, 523–531. [Google Scholar]
- Burgess, N.K.; Dao, T.P.; Stanley, A.M.; Fleming, K.G. β-barrel proteins that reside in the Escherichia coli outer membrane in vivo demonstrate varied folding behavior in vitro. J. Biol. Chem 2008, 283, 26748–26758. [Google Scholar]
- Hagan, C.L.; Silhavy, T.J.; Kahne, D. β-Barrel membrane protein assembly by the Bam complex. Annu. Rev. Biochem 2011, 80, 189–210. [Google Scholar]
- Hagan, C.L.; Kim, S.; Kahne, D. Reconstitution of outer membrane protein assembly from purified components. Science 2010, 328, 890–892. [Google Scholar]
- Chacinska, A.; Koehler, C.M.; Milenkovic, D.; Lithgow, T.; Pfanner, N. Importing mitochondrial proteins: Machineries and mechanisms. Cell 2009, 138, 628–644. [Google Scholar]
- Hoppins, S.C.; Nargang, F.E. The Tim8-Tim13 complex of Neurospora crassa functions in the assembly of proteins into both mitochondrial membranes. J. Biol. Chem 2004, 279, 12396–12405. [Google Scholar]
- Wiedemann, N.; Truscott, K.N.; Pfannschmidt, S.; Guiard, B.; Meisinger, C.; Pfanner, N. Biogenesis of the protein import channel Tom40 of the mitochondrial outer membrane: Intermembrane space components are involved in an early stage of the assembly pathway. J. Biol. Chem 2004, 279, 18188–18194. [Google Scholar]
- Wiedemann, N.; Kozjak, V.; Chacinska, A.; Schonfisch, B.; Rospert, S.; Ryan, M.T.; Pfanner, N.; Meisinger, C. Machinery for protein sorting and assembly in the mitochondrial outer membrane. Nature 2003, 424, 565–571. [Google Scholar]
- Paschen, S.A.; Waizenegger, T.; Stan, T.; Preuss, M.; Cyrklaff, M.; Hell, K.; Rapaport, D.; Neupert, W. Evolutionary conservation of biogenesis of beta-barrel membrane proteins. Nature 2003, 426, 862–866. [Google Scholar]
- Gentle, I.; Gabriel, K.; Beech, P.; Waller, R.; Lithgow, T. The Omp85 family of proteins is essential for outer membrane biogenesis in mitochondria and bacteria. J. Cell Biol 2004, 164, 19–24. [Google Scholar]
- Kozjak, V.; Wiedemann, N.; Milenkovic, D.; Lohaus, C.; Meyer, H.E.; Guiard, B.; Meisinger, C.; Pfanner, N. An essential role of Sam50 in the protein sorting and assembly machinery of the mitochondrial outer membrane. J. Biol. Chem 2003, 278, 48520–48523. [Google Scholar]
- Voulhoux, R.; Bos, M.P.; Geurtsen, J.; Mols, M.; Tommassen, J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science 2003, 299, 262–265. [Google Scholar]
- Reumann, S.; Davila-Aponte, J.; Keegstra, K. The evolutionary origin of the protein-translocating channel of chloroplastic envelope membranes: Identification of a cyanobacterial homolog. Proc. Natl. Acad. Sci. USA 1999, 96, 784–789. [Google Scholar]
- Wu, T.; Malinverni, J.; Ruiz, N.; Kim, S.; Silhavy, T.J.; Kahne, D. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell 2005, 121, 235–245. [Google Scholar]
- Werner, J.; Misra, R. YaeT (Omp85) affects the assembly of lipid-dependent and lipid-independent outer membrane proteins of Escherichia coli. Mol. Microbiol 2005, 57, 1450–1459. [Google Scholar]
- Doerrler, W.T.; Raetz, C.R. Loss of outer membrane proteins without inhibition of lipid export in an Escherichia coli YaeT mutant. J. Biol. Chem 2005, 280, 27679–27687. [Google Scholar]
- Sanchez-Pulido, L.; Devos, D.; Genevrois, S.; Vicente, M.; Valencia, A. POTRA: A conserved domain in the FtsQ family and a class of beta-barrel outer membrane proteins. Trends Biochem. Sci 2003, 28, 523–526. [Google Scholar]
- Kim, S.; Malinverni, J.C.; Sliz, P.; Silhavy, T.J.; Harrison, S.C.; Kahne, D. Structure and function of an essential component of the outer membrane protein assembly machine. Science 2007, 317, 961–964. [Google Scholar]
- Bos, M.P.; Robert, V.; Tommassen, J. Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep 2007, 8, 1149–1154. [Google Scholar]
- Genevrois, S.; Steeghs, L.; Roholl, P.; Letesson, J.J.; van der Ley, P. The Omp85 protein of Neisseria meningitidis is required for lipid export to the outer membrane. EMBO J 2003, 22, 1780–1789. [Google Scholar]
- Chimalakonda, G.; Ruiz, N.; Chng, S.S.; Garner, R.A.; Kahne, D.; Silhavy, T.J. Lipoprotein LptE is required for the assembly of LptD by the beta-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. USA 2011, 108, 2492–2497. [Google Scholar]
- Malinverni, J.C.; Werner, J.; Kim, S.; Sklar, J.G.; Kahne, D.; Misra, R.; Silhavy, T.J. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol. Microbiol 2006, 61, 151–164. [Google Scholar]
- Sklar, J.G.; Wu, T.; Gronenberg, L.S.; Malinverni, J.C.; Kahne, D.; Silhavy, T.J. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 2007, 104, 6400–6405. [Google Scholar]
- Hagan, C.L.; Silhavy, T.J.; Kahne, D.E. β-Barrel Membrane Protein Assembly by the Bam Complex. Annu. Rev. Biochem 2011, 80, 189–210. [Google Scholar]
- Ieva, R.; Bernstein, H.D. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc. Natl. Acad. Sci. USA 2009, 106, 19120–19125. [Google Scholar]
- Ieva, R.; Tian, P.; Peterson, J.H.; Bernstein, H.D. Sequential and spatially restricted interactions of assembly factors with an autotransporter beta domain. Proc. Natl. Acad. Sci. USA 2011, 108, E383–E391. [Google Scholar]
- Walther, D.M.; Rapaport, D.; Tommassen, J. Biogenesis of β-barrel membrane proteins in bacteria and eukaryotes: Evolutionary conservation and divergence. Cell. Mol. Life Sci 2009, 66, 2789–2804. [Google Scholar]
- Gatsos, X.; Perry, A.J.; Anwari, K.; Dolezal, P.; Wolynec, P.P.; Likic, V.A.; Purcell, A.W.; Buchanan, S.K.; Lithgow, T. Protein secretion and outer membrane assembly in Alphaproteobacteria. FEMS Microbiol. Rev 2008, 32, 995–1009. [Google Scholar]
- Habib, S.J.; Waizenegger, T.; Niewienda, A.; Paschen, S.A.; Neupert, W.; Rapaport, D. The N-terminal domain of Tob55 has a receptor-like function in the biogenesis of mitochondrial β-barrel proteins. J. Cell Biol 2007, 176, 77–88. [Google Scholar]
- Stroud, D.A.; Becker, T.; Qiu, J.; Stojanovski, D.; Pfannschmidt, S.; Wirth, C.; Hunte, C.; Guiard, B.; Meisinger, C.; Pfanner, N.; et al. Biogenesis of mitochondrial beta-barrel proteins: The POTRA domain is involved in precursor release from the SAM complex. Mol. Biol. Cell 2011, 22, 2823–2833. [Google Scholar]
- Kutik, S.; Stojanovski, D.; Becker, L.; Becker, T.; Meinecke, M.; Kruger, V.; Prinz, C.; Meisinger, C.; Guiard, B.; Wagner, R.; et al. Dissecting membrane insertion of mitochondrial β-barrel proteins. Cell 2008, 132, 1011–1024. [Google Scholar]
- Meisinger, C.; Rissler, M.; Chacinska, A.; Szklarz, L.K.; Milenkovic, D.; Kozjak, V.; Schonfisch, B.; Lohaus, C.; Meyer, H.E.; Yaffe, M.P.; et al. The mitochondrial morphology protein Mdm10 functions in assembly of the preprotein translocase of the outer membrane. Dev. Cell 2004, 7, 61–71. [Google Scholar]
- Meisinger, C.; Pfannschmidt, S.; Rissler, M.; Milenkovic, D.; Becker, T.; Stojanovski, D.; Youngman, M.J.; Jensen, R.E.; Chacinska, A.; Guiard, B.; et al. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria. EMBO J 2007, 26, 2229–2239. [Google Scholar]
- Chan, N.C.; Lithgow, T. The peripheral membrane subunits of the SAM complex function codependently in mitochondrial outer membrane biogenesis. Mol. Biol. Cell 2008, 19, 126–136. [Google Scholar]
- Kornmann, B.; Currie, E.; Collins, S.R.; Schuldiner, M.; Nunnari, J.; Weissman, J.S.; Walter, P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science 2009, 325, 477–481. [Google Scholar]
- Thornton, N.; Stroud, D.A.; Milenkovic, D.; Guiard, B.; Pfanner, N.; Becker, T. Two modular forms of the mitochondrial sorting and assembly machinery are involved in biogenesis of α-helical outer membrane proteins. J. Mol. Biol 2010, 396, 540–549. [Google Scholar]
- Wiedemann, N.; Meisinger, C.; Pfanner, N. Cell biology. Connecting organelles. Science 2009, 325, 403–404. [Google Scholar]
- Kozjak-Pavlovic, V.; Ross, K.; Benlasfer, N.; Kimmig, S.; Karlas, A.; Rudel, T. Conserved roles of Sam50 and metaxins in VDAC biogenesis. EMBO Rep 2007, 8, 576–582. [Google Scholar]
- Hill, K.; Model, K.; Ryan, M.T.; Dietmeier, K.; Martin, F.; Wagner, R.; Pfanner, N. Tom40 forms the hydrophilic channel of the mitochondrial import pore for preproteins [see comment]. Nature 1998, 395, 516–521. [Google Scholar]
- Meisinger, C.; Ryan, M.T.; Hill, K.; Model, K.; Lim, J.H.; Sickmann, A.; Muller, H.; Meyer, H.E.; Wagner, R.; Pfanner, N. Protein import channel of the outer mitochondrial membrane: A highly stable Tom40-Tom22 core structure differentially interacts with preproteins, small tom proteins, and import receptors. Mol. Cell. Biol 2001, 21, 2337–2348. [Google Scholar]
- Model, K.; Meisinger, C.; Prinz, T.; Wiedemann, N.; Truscott, K.N.; Pfanner, N.; Ryan, M.T. Multistep assembly of the protein import channel of the mitochondrial outer membrane. Nat. Struct. Biol 2001, 8, 361–370. [Google Scholar]
- Model, K.; Prinz, T.; Ruiz, T.; Radermacher, M.; Krimmer, T.; Kuhlbrandt, W.; Pfanner, N.; Meisinger, C. Protein translocase of the outer mitochondrial membrane: Role of import receptors in the structural organization of the TOM complex. J. Mol. Biol 2002, 316, 657–666. [Google Scholar]
- Wideman, J.G.; Go, N.E.; Klein, A.; Redmond, E.; Lackey, S.W.; Tao, T.; Kalbacher, H.; Rapaport, D.; Neupert, W.; Nargang, F.E. Roles of the Mdm10, Tom7, Mdm12, and Mmm1 proteins in the assembly of mitochondrial outer membrane proteins in Neurospora crassa. Mol. Biol. Cell 2010, 21, 1725–1736. [Google Scholar]
- Becker, T.; Wenz, L.S.; Thornton, N.; Stroud, D.; Meisinger, C.; Wiedemann, N.; Pfanner, N. Biogenesis of mitochondria: Dual role of Tom7 in modulating assembly of the preprotein translocase of the outer membrane. J. Mol. Biol 2011, 405, 113–124. [Google Scholar]
- Yamano, K.; Tanaka-Yamano, S.; Endo, T. Mdm10 as a dynamic constituent of the TOB/SAM complex directs coordinated assembly of Tom40. EMBO Rep 2010, 11, 187–193. [Google Scholar]
- Yamano, K.; Tanaka-Yamano, S.; Endo, T. Tom7 regulates Mdm10-mediated assembly of the mitochondrial import channel protein Tom40. J. Biol. Chem 2010, 285, 41222–41231. [Google Scholar]
- Kornmann, B. ERMES, a multifunctional complex connecting endoplasmic reticulum and mitochondria. Med. Sci. (Paris) 2010, 26, 145–146. [Google Scholar]
- Becker, T.; Pfannschmidt, S.; Guiard, B.; Stojanovski, D.; Milenkovic, D.; Kutik, S.; Pfanner, N.; Meisinger, C.; Wiedemann, N. Biogenesis of the mitochondrial TOM complex: Mim1 promotes insertion and assembly of signal-anchored receptors. J. Biol. Chem 2008, 283, 120–127. [Google Scholar]
- Hulett, J.M.; Lueder, F.; Chan, N.C.; Perry, A.J.; Wolynec, P.; Likic, V.A.; Gooley, P.R.; Lithgow, T. The transmembrane segment of Tom20 is recognized by Mim1 for docking to the mitochondrial TOM complex. J. Mol. Biol 2008, 376, 694–704. [Google Scholar]
- Ott, C.; Ross, K.; Straub, S.; Thiede, B.; Gotz, M.; Goosmann, C.; Krischke, M.; Mueller, M.J.; Krohne, G.; Rudel, T.; et al. Sam50 functions in mitochondrial intermembrane space bridging and biogenesis of respiratory complexes. Mol. Cell. Biol 2012, 32, 1173–1188. [Google Scholar]
- Walther, D.M.; Papic, D.; Bos, M.P.; Tommassen, J.; Rapaport, D. Signals in bacterial β-barrel proteins are functional in eukaryotic cells for targeting to and assembly in mitochondria. Proc. Natl. Acad. Sci. USA 2009, 106, 2531–2536. [Google Scholar]
- Kozjak-Pavlovic, V.; Ott, C.; Gotz, M.; Rudel, T. Neisserial Omp85 protein is selectively recognized and assembled into functional complexes in the outer membrane of human mitochondria. J. Biol. Chem 2011, 286, 27019–27026. [Google Scholar]
- Muller, J.E.; Papic, D.; Ulrich, T.; Grin, I.; Schutz, M.; Oberhettinger, P.; Tommassen, J.; Linke, D.; Dimmer, K.S.; Autenrieth, I.B.; et al. Mitochondria can recognize and assemble fragments of a beta-barrel structure. Mol. Biol. Cell 2011, 22, 1638–1647. [Google Scholar]
- Jiang, J.H.; Davies, J.K.; Lithgow, T.; Strugnell, R.A.; Gabriel, K. Targeting of Neisserial PorB to the mitochondrial outer membrane: An insight on the evolution of β-barrel protein assembly machines. Mol. Microbiol 2011, 82, 976–987. [Google Scholar]
- Struyve, M.; Moons, M.; Tommassen, J. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol 1991, 218, 141–148. [Google Scholar]
- Kozjak-Pavlovic, V.; Dian-Lothrop, E.A.; Meinecke, M.; Kepp, O.; Ross, K.; Rajalingam, K.; Harsman, A.; Hauf, E.; Brinkmann, V.; Gunther, D.; et al. Bacterial porin disrupts mitochondrial membrane potential and sensitizes host cells to apoptosis. PLoS Pathog 2009, 5. [Google Scholar] [CrossRef]
- Muller, A.; Gunther, D.; Brinkmann, V.; Hurwitz, R.; Meyer, T.F.; Rudel, T. Targeting of the pro-apoptotic VDAC-like porin (PorB) of Neisseria gonorrhoeae to mitochondria of infected cells. EMBO J 2000, 19, 5332–5343. [Google Scholar]
- Muller, A.; Rassow, J.; Grimm, J.; Machuy, N.; Meyer, T.F.; Rudel, T. VDAC and the bacterial porin PorB of Neisseria gonorrhoeae share mitochondrial import pathways. EMBO J 2002, 21, 1916–1929. [Google Scholar]
- Massari, P.; Ho, Y.; Wetzler, L.M. Neisseria meningitidis porin PorB interacts with mitochondria and protects cells from apoptosis. Proc. Natl. Acad. Sci. USA 2000, 97, 9070–9075. [Google Scholar]
- Massari, P.; King, C.A.; Ho, A.Y.; Wetzler, L.M. Neisserial PorB is translocated to the mitochondria of HeLa cells infected with Neisseria meningitidis and protects cells from apoptosis. Cell. Microbiol 2003, 5, 99–109. [Google Scholar]
- Choi, C.H.; Lee, E.Y.; Lee, Y.C.; Park, T.I.; Kim, H.J.; Hyun, S.H.; Kim, S.A.; Lee, S.K.; Lee, J.C. Outer membrane protein 38 of Acinetobacter baumannii localizes to the mitochondria and induces apoptosis of epithelial cells. Cell. Microbiol 2005, 7, 1127–1138. [Google Scholar]
- Genestier, A.L.; Michallet, M.C.; Prevost, G.; Bellot, G.; Chalabreysse, L.; Peyrol, S.; Thivolet, F.; Etienne, J.; Lina, G.; Vallette, F.M.; et al. Staphylococcus aureus Panton-Valentine leukocidin directly targets mitochondria and induces Bax-independent apoptosis of human neutrophils. J. Clin. Invest 2005, 115, 3117–3127. [Google Scholar]
- Walther, D.M.; Bos, M.P.; Rapaport, D.; Tommassen, J. The mitochondrial porin, VDAC, has retained the ability to be assembled in the bacterial outer membrane. Mol. Biol. Evol 2010, 27, 887–895. [Google Scholar]
- Danial, N.N.; Korsmeyer, S.J. Cell death: Critical control points. Cell 2004, 116, 205–219. [Google Scholar]
- Rudel, T.; Kepp, O.; Kozjak-Pavlovic, V. Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat. Rev. Microbiol 2010, 8, 693–705. [Google Scholar]
- Jiang, J.H.; Tong, J.; Gabriel, K. Hijacking mitochondria: Bacterial toxins that modulate mitochondrial function. IUBMB Life 2012, 64, 397–401. [Google Scholar]
- Tanabe, M.; Nimigean, C.M.; Iverson, T.M. Structural basis for solute transport, nucleotide regulation, and immunological recognition of Neisseria meningitidis PorB. Proc. Natl. Acad. Sci. USA 2010, 107, 6811–6816. [Google Scholar]
- Rudel, T.; Schmid, A.; Benz, R.; Kolb, H.A.; Lang, F.; Meyer, T.F. Modulation of Neisseria porin (PorB) by cytosolic ATP/GTP of target cells: Parallels between pathogen accommodation and mitochondrial endosymbiosis. Cell 1996, 85, 391–402. [Google Scholar]
- Park, J.S.; Lee, W.C.; Yeo, K.J.; Ryu, K.S.; Kumarasiri, M.; Hesek, D.; Lee, M.; Mobashery, S.; Song, J.H.; Kim, S.I.; et al. Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J 2012, 26, 219–228. [Google Scholar]
- Kaneko, J.; Kimura, T.; Narita, S.; Tomita, T.; Kamio, Y. Complete nucleotide sequence and molecular characterization of the temperate staphylococcal bacteriophage phiPVL carrying Panton-Valentine leukocidin genes. Gene 1998, 215, 57–67. [Google Scholar]
- Yamashita, K.; Kawai, Y.; Tanaka, Y.; Hirano, N.; Kaneko, J.; Tomita, N.; Ohta, M.; Kamio, Y.; Yao, M.; Tanaka, I. Crystal structure of the octameric pore of staphylococcal gamma-hemolysin reveals the β-barrel pore formation mechanism by two components. Proc. Natl. Acad. Sci. USA 2011, 108, 17314–17319. [Google Scholar]
- Roblin, P.; Guillet, V.; Joubert, O.; Keller, D.; Erard, M.; Maveyraud, L.; Prevost, G.; Mourey, L. A covalent S-F heterodimer of leucotoxin reveals molecular plasticity of β-barrel pore-forming toxins. Proteins 2008, 71, 485–496. [Google Scholar]

| Bacterial species | β-barrel protein | References |
|---|---|---|
| Bacterial βbarrels target to mitochondria during infection | ||
| Neisseria gonorrhoeae | PorB | [58,60,62–64] |
| Neisseria meningitidis | PorB | [60,65,66] |
| Acinetobacter baumannii | Omp38 (AbOmpA) | [67] |
| Staphylococcus aureus | Panton-Valentine leukocidin (PVL) | [68] |
| Bacterial β-barrels tested to be expressed in mitochondria | ||
| E. coli | PhoE, OmpA, OmpC | [57,58] |
| Yersinia enterocolitica | YadA | [59] |
| Neisseria meningitidis | Omp85(BamA) | [57] |
| Neisseria gonorrhoeae | Omp85(BamA) | [58] |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Jiang, J.-H.; Tong, J.; Tan, K.S.; Gabriel, K. From Evolution to Pathogenesis: The Link Between β-Barrel Assembly Machineries in the Outer Membrane of Mitochondria and Gram-Negative Bacteria. Int. J. Mol. Sci. 2012, 13, 8038-8050. https://doi.org/10.3390/ijms13078038
Jiang J-H, Tong J, Tan KS, Gabriel K. From Evolution to Pathogenesis: The Link Between β-Barrel Assembly Machineries in the Outer Membrane of Mitochondria and Gram-Negative Bacteria. International Journal of Molecular Sciences. 2012; 13(7):8038-8050. https://doi.org/10.3390/ijms13078038
Chicago/Turabian StyleJiang, Jhih-Hang, Janette Tong, Kher Shing Tan, and Kipros Gabriel. 2012. "From Evolution to Pathogenesis: The Link Between β-Barrel Assembly Machineries in the Outer Membrane of Mitochondria and Gram-Negative Bacteria" International Journal of Molecular Sciences 13, no. 7: 8038-8050. https://doi.org/10.3390/ijms13078038
APA StyleJiang, J. -H., Tong, J., Tan, K. S., & Gabriel, K. (2012). From Evolution to Pathogenesis: The Link Between β-Barrel Assembly Machineries in the Outer Membrane of Mitochondria and Gram-Negative Bacteria. International Journal of Molecular Sciences, 13(7), 8038-8050. https://doi.org/10.3390/ijms13078038