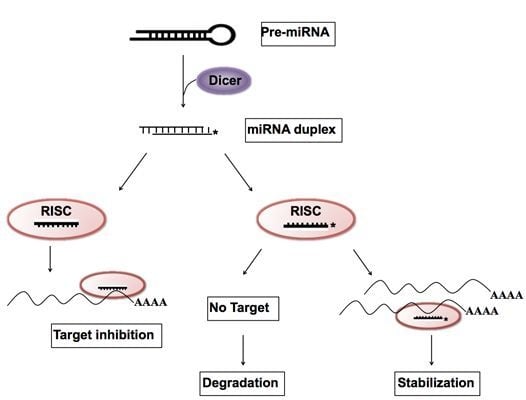
Up-Regulation of microRNA* Strands by Their Target Transcripts
Abstract
:1. Introduction
2. Results and Discussion
3. Experimental Section
3.1. Cell Culture
3.2. Construction of Artificial miRNA Target
3.3. Luciferase Assays
3.4. Isolation of miRNA and Quantitative Real-Time PCR (qRT-PCR) Analysis
3.5. Immunoprecipitation
3.6. Isolation of Ago2-Associated miRNA
3.7. Northern Blot Analysis for miRNA
3.8. Statistical Analysis
4. Conclusions
Acknowledgments
Conflict of Interest
References
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Kim, Y.K.; Heo, I.; Kim, V.N. Modifications of small RNAs and their associated proteins. Cell 2010, 143, 703–709. [Google Scholar]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar]
- Bushati, N.; Cohen, S.M. microRNA Functions. Annu. Rev. Cell Dev. Biol 2007, 23, 175–205. [Google Scholar]
- Lee, Y.; Jeon, K.; Lee, J.T.; Kim, S.; Kim, V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 2002, 21, 4663–4670. [Google Scholar]
- Rand, T.A.; Petersen, S.; Du, F.; Wang, X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 2005, 123, 621–629. [Google Scholar]
- Matranga, C.; Tomari, Y.; Shin, C.; Bartel, D.P.; Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 2005, 123, 607–620. [Google Scholar]
- Wang, H.W.; Noland, C.; Siridechadilok, B.; Taylor, D.W.; Ma, E.; Felderer, K.; Doudna, J.A.; Nogales, E. Structural insights into RNA processing by the human RISC-loading complex. Nat. Struct. Mol. Biol 2009, 16, 1148–1153. [Google Scholar]
- Leuschner, P.J.; Ameres, S.L.; Kueng, S.; Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep 2006, 7, 314–320. [Google Scholar]
- Iwasaki, S.; Kawamata, T.; Tomari, Y. Drosophila argonaute1 and argonaute2 employ distinct mechanisms for translational repression. Mol. Cell 2009, 34, 58–67. [Google Scholar]
- Diederichs, S.; Haber, D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 2007, 131, 1097–1108. [Google Scholar]
- Bhayani, M.K.; Calin, G.A.; Lai, S.Y. Functional relevance of miRNA sequences in human disease. Mutat. Res 2012, 731, 14–19. [Google Scholar]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell 2003, 115, 209–216. [Google Scholar]
- Schwarz, D.S.; Hutvagner, G.; Du, T.; Xu, Z.; Aronin, N.; Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell 2003, 115, 199–208. [Google Scholar]
- Griffiths-Jones, S.; Hui, J.H.; Marco, A.; Ronshaugen, M. MicroRNA evolution by arm switching. EMBO Rep 2011, 12, 172–177. [Google Scholar]
- Li, S.C.; Liao, Y.L.; Ho, M.R.; Tsai, K.W.; Lai, C.H.; Lin, W.C. miRNA arm selection and isomiR distribution in gastric cancer. BMC Genomics Electron. Resour 2012, 13, S13. [Google Scholar]
- Okamura, K.; Phillips, M.D.; Tyler, D.M.; Duan, H.; Chou, Y.T.; Lai, E.C. The regulatory activity of microRNA* species has substantial influence on microRNA and 3′ UTR evolution. Nat. Struct. Mol. Biol 2008, 15, 354–363. [Google Scholar]
- Arvey, A.; Larsson, E.; Sander, C.; Leslie, C.S.; Marks, D.S. Target mRNA abundance dilutes microRNA and siRNA activity. Mol. Syst. Biol 2010, 6, 363. [Google Scholar]
- Larsson, E.; Sander, C.; Marks, D. mRNA turnover rate limits siRNA and microRNA efficacy. Mol. Syst. Biol 2010, 6, 433. [Google Scholar]
- Chatterjee, S.; Fasler, M.; Bussing, I.; Grosshans, H. Target-mediated protection of endogenous microRNAs in C. elegans. Dev. Cell 2011, 20, 388–396. [Google Scholar]
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726. [Google Scholar]
- Lee, H.J.; Palkovits, M.; Young, W.S., 3rd. miR-7b, a microRNA up-regulated in the hypothalamus after chronic hyperosmolar stimulation, inhibits Fos translation. Proc. Natl. Acad. Sci. USA 2006, 103, 15669–15674. [Google Scholar]
- Hutvagner, G.; Simard, M.J. Argonaute proteins: Key players in RNA silencing. Nat. Rev. Mol. Cell Biol 2008, 9, 22–32. [Google Scholar]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar]
- Liu, J.; Carmell, M.A.; Rivas, F.V.; Marsden, C.G.; Thomson, J.M.; Song, J.J.; Hammond, S.M.; Joshua-Tor, L.; Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science 2004, 305, 1437–1441. [Google Scholar]
- Kwak, P.B.; Tomari, Y. The N domain of Argonaute drives duplex unwinding during RISC assembly. Nat. Struct. Mol. Biol 2012, 19, 145–151. [Google Scholar]







© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kang, S.-M.; Choi, J.-W.; Hong, S.-H.; Lee, H.-J. Up-Regulation of microRNA* Strands by Their Target Transcripts. Int. J. Mol. Sci. 2013, 14, 13231-13240. https://doi.org/10.3390/ijms140713231
Kang S-M, Choi J-W, Hong S-H, Lee H-J. Up-Regulation of microRNA* Strands by Their Target Transcripts. International Journal of Molecular Sciences. 2013; 14(7):13231-13240. https://doi.org/10.3390/ijms140713231
Chicago/Turabian StyleKang, Sung-Min, Ji-Woong Choi, Su-Hyung Hong, and Heon-Jin Lee. 2013. "Up-Regulation of microRNA* Strands by Their Target Transcripts" International Journal of Molecular Sciences 14, no. 7: 13231-13240. https://doi.org/10.3390/ijms140713231
APA StyleKang, S. -M., Choi, J. -W., Hong, S. -H., & Lee, H. -J. (2013). Up-Regulation of microRNA* Strands by Their Target Transcripts. International Journal of Molecular Sciences, 14(7), 13231-13240. https://doi.org/10.3390/ijms140713231