Long Non-Coding RNAs in Haematological Malignancies
Abstract
:1. Introduction
2. Classes of lncRNAs and lncRNA Functionality
2.1. Classification by Genomic Location
2.2. Classification by Specific Characteristics
2.3. Classification as cis or trans-Acting Molecules
2.4. lncRNA Functionality
3. LncRNAs Deregulated in Haematological Malignancies
3.1. Host Genes of Small RNAs
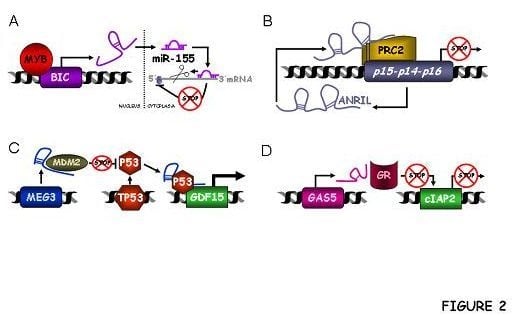
3.1.1. BIC and C13ORF25
3.1.2. nc886 or vtRNA2-1
3.1.3. PVT1
3.2. LncRNAs with Oncogenic Properties
ANRIL or CDKN2B-AS1
3.3. LncRNAs with Tumor Suppressor Properties
3.3.1. MEG3
3.3.2. DLEU1 and DLEU2
3.3.3. GAS5
3.4. LncRNAs with Dual Functions
3.4.1. H19
3.4.2. T-UCRs
3.5. LncRNAs Poorly Characterized in Haematological Malignancies
4. LncRNAs Involved in Hematopoiesis
5. Regulation of the Expression of lncRNAs Involved in Haematological Malignancies
6. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Mattick, J.S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep 2001, 2, 986–991. [Google Scholar]
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar]
- Lin, M.F.; Carlson, J.W.; Crosby, M.A.; Matthews, B.B.; Yu, C.; Park, S.; Wan, K.H.; Schroeder, A.J.; Gramates, L.S.; St Pierre, S.E.; et al. Revisiting the protein-coding gene catalog of Drosophila melanogaster using 12 fly genomes. Genome Res 2007, 17, 1823–1836. [Google Scholar]
- Lin, M.F.; Deoras, A.N.; Rasmussen, M.D.; Kellis, M. Performance and scalability of discriminative metrics for comparative gene identification in 12 Drosophila genomes. PLoS Comput. Biol 2008, 4, e1000067. [Google Scholar]
- Pueyo, J.I.; Couso, J.P. Tarsal-less peptides control Notch signalling through the Shavenbaby transcription factor. Dev. Biol 2011, 355, 183–193. [Google Scholar]
- Ingolia, N.T.; Lareau, L.F.; Weissman, J.S. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell 2011, 147, 789–802. [Google Scholar]
- Banfai, B.; Jia, H.; Khatun, J.; Wood, E.; Risk, B.; Gundling, W.E., Jr; Kundaje, A.; Gunawardena, H.P.; Yu, Y.; Xie, L.; et al. Long noncoding RNAs are rarely translated in two human cell lines. Genome Res. 2012, 22, 1646–1657. [Google Scholar]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res 2012, 22, 1775–1789. [Google Scholar]
- Kloc, M.; Wilk, K.; Vargas, D.; Shirato, Y.; Bilinski, S.; Etkin, L.D. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development 2005, 132, 3445–3457. [Google Scholar]
- Wadler, C.S.; Vanderpool, C.K. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. USA 2007, 104, 20454–20459. [Google Scholar]
- Dinger, M.E.; Pang, K.C.; Mercer, T.R.; Mattick, J.S. Differentiating protein-coding and noncoding RNA: Challenges and ambiguities. PLoS Comput. Biol 2008, 4, e1000176. [Google Scholar]
- Leygue, E. Steroid receptor RNA activator (SRA1): Unusual bifaceted gene products with suspected relevance to breast cancer. Nucl. Recept. Signal 2007, 5, e006. [Google Scholar]
- Jenny, A.; Hachet, O.; Zavorszky, P.; Cyrklaff, A.; Weston, M.D.; Johnston, D.S.; Erdelyi, M.; Ephrussi, A. A translation-independent role of oskar RNA in early Drosophila oogenesis. Development 2006, 133, 2827–2833. [Google Scholar]
- Candeias, M.M.; Malbert-Colas, L.; Powell, D.J.; Daskalogianni, C.; Maslon, M.M.; Naski, N.; Bourougaa, K.; Calvo, F.; Fahraeus, R. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat. Cell Biol 2008, 10, 1098–1105. [Google Scholar]
- Guttman, M.; Amit, I.; Garber, M.; French, C.; Lin, M.F.; Feldser, D.; Huarte, M.; Zuk, O.; Carey, B.W.; Cassady, J.P.; et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009, 458, 223–227. [Google Scholar]
- Tilgner, H.; Knowles, D.G.; Johnson, R.; Davis, C.A.; Chakrabortty, S.; Djebali, S.; Curado, J.; Snyder, M.; Gingeras, T.R.; Guigo, R. Deep sequencing of subcellular RNA fractions shows splicing to be predominantly co-transcriptional in the human genome but inefficient for lncRNAs. Genome Res 2012, 22, 1616–1625. [Google Scholar]
- Gibb, E.A.; Vucic, E.A.; Enfield, K.S.; Stewart, G.L.; Lonergan, K.M.; Kennett, J.Y.; Becker-Santos, D.D.; MacAulay, C.E.; Lam, S.; Brown, C.J.; et al. Human cancer long non-coding RNA transcriptomes. PLoS One 2011, 6, e25915. [Google Scholar]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar]
- Hauptman, N.; Glavac, D. Long non-coding RNA in cancer. Int. J. Mol. Sci 2013, 14, 4655–4669. [Google Scholar]
- Agirre, X.; Jimenez-Velasco, A.; San Jose-Eneriz, E.; Garate, L.; Bandres, E.; Cordeu, L.; Aparicio, O.; Saez, B.; Navarro, G.; Vilas-Zornoza, A.; et al. Down-regulation of hsa-miR-10a in chronic myeloid leukemia CD34+ cells increases USF2-mediated cell growth. Mol. Cancer Res 2008, 6, 1830–1840. [Google Scholar]
- Agirre, X.; Vilas-Zornoza, A.; Jimenez-Velasco, A.; Martin-Subero, J.I.; Cordeu, L.; Garate, L.; San Jose-Eneriz, E.; Abizanda, G.; Rodriguez-Otero, P.; Fortes, P.; et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res 2009, 69, 4443–4453. [Google Scholar]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar]
- Katayama, S.; Tomaru, Y.; Kasukawa, T.; Waki, K.; Nakanishi, M.; Nakamura, M.; Nishida, H.; Yap, C.C.; Suzuki, M.; Kawai, J.; et al. Antisense transcription in the mammalian transcriptome. Science 2005, 309, 1564–1566. [Google Scholar]
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar]
- Galante, P.A.; Vidal, D.O.; de Souza, J.E.; Camargo, A.A.; de Souza, S.J. Sense-antisense pairs in mammals: Functional and evolutionary considerations. Genome Biol 2007, 8, R40. [Google Scholar]
- Yap, K.L.; Li, S.; Munoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar]
- Krystal, G.W.; Armstrong, B.C.; Battey, J.F. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol. Cell Biol 1990, 10, 4180–4191. [Google Scholar]
- Hastings, M.L.; Milcarek, C.; Martincic, K.; Peterson, M.L.; Munroe, S.H. Expression of the thyroid hormone receptor gene, erbAalpha, in B lymphocytes: Alternative mRNA processing is independent of differentiation but correlates with antisense RNA levels. Nucleic Acids Res 1997, 25, 4296–4300. [Google Scholar]
- Yan, M.D.; Hong, C.C.; Lai, G.M.; Cheng, A.L.; Lin, Y.W.; Chuang, S.E. Identification and characterization of a novel gene Saf transcribed from the opposite strand of Fas. Hum. Mol. Genet 2005, 14, 1465–1474. [Google Scholar]
- Annilo, T.; Kepp, K.; Laan, M. Natural antisense transcript of natriuretic peptide precursor A (NPPA): Structural organization and modulation of NPPA expression. BMC Mol. Biol 2009, 10, 81. [Google Scholar]
- Allo, M.; Buggiano, V.; Fededa, J.P.; Petrillo, E.; Schor, I.; de la Mata, M.; Agirre, E.; Plass, M.; Eyras, E.; Elela, S.A.; et al. Control of alternative splicing through siRNA-mediated transcriptional gene silencing. Nat. Struct. Mol. Biol 2009, 16, 717–724. [Google Scholar]
- Faghihi, M.A.; Modarresi, F.; Khalil, A.M.; Wood, D.E.; Sahagan, B.G.; Morgan, T.E.; Finch, C.E.; St Laurent, G., 3rd; Kenny, P.J.; Wahlestedt, C. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat. Med. 2008, 14, 723–730. [Google Scholar]
- Guil, S.; Esteller, M. cis-acting noncoding RNAs: Friends and foes. Nat. Struct. Mol. Biol 2012, 19, 1068–1075. [Google Scholar]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; Ching, K.A.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet 2007, 39, 311–318. [Google Scholar]
- Heintzman, N.D.; Hon, G.C.; Hawkins, R.D.; Kheradpour, P.; Stark, A.; Harp, L.F.; Ye, Z.; Lee, L.K.; Stuart, R.K.; Ching, C.W.; et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 2009, 459, 108–112. [Google Scholar] [Green Version]
- Visel, A.; Blow, M.J.; Li, Z.; Zhang, T.; Akiyama, J.A.; Holt, A.; Plajzer-Frick, I.; Shoukry, M.; Wright, C.; Chen, F.; et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 2009, 457, 854–858. [Google Scholar]
- Kim, T.K.; Hemberg, M.; Gray, J.M.; Costa, A.M.; Bear, D.M.; Wu, J.; Harmin, D.A.; Laptewicz, M.; Barbara-Haley, K.; Kuersten, S.; et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010, 465, 182–187. [Google Scholar]
- Wang, D.; Garcia-Bassets, I.; Benner, C.; Li, W.; Su, X.; Zhou, Y.; Qiu, J.; Liu, W.; Kaikkonen, M.U.; Ohgi, K.A.; et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 2011, 474, 390–394. [Google Scholar]
- De Santa, F.; Barozzi, I.; Mietton, F.; Ghisletti, S.; Polletti, S.; Tusi, B.K.; Muller, H.; Ragoussis, J.; Wei, C.L.; Natoli, G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 2010, 8, e1000384. [Google Scholar]
- Orom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar]
- Melo, C.A.; Drost, J.; Wijchers, P.J.; van de Werken, H.; de Wit, E.; Oude Vrielink, J.A.; Elkon, R.; Melo, S.A.; Leveille, N.; Kalluri, R.; et al. eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol. Cell 2013, 49, 524–535. [Google Scholar]
- Lai, F.; Orom, U.A.; Cesaroni, M.; Beringer, M.; Taatjes, D.J.; Blobel, G.A.; Shiekhattar, R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013, 494, 497–501. [Google Scholar]
- Bejerano, G.; Pheasant, M.; Makunin, I.; Stephen, S.; Kent, W.J.; Mattick, J.S.; Haussler, D. Ultraconserved elements in the human genome. Science 2004, 304, 1321–1325. [Google Scholar]
- Ahituv, N.; Zhu, Y.; Visel, A.; Holt, A.; Afzal, V.; Pennacchio, L.A.; Rubin, E.M. Deletion of ultraconserved elements yields viable mice. PLoS Biol 2007, 5, e234. [Google Scholar]
- Calin, G.A.; Liu, C.G.; Ferracin, M.; Hyslop, T.; Spizzo, R.; Sevignani, C.; Fabbri, M.; Cimmino, A.; Lee, E.J.; Wojcik, S.E.; et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell 2007, 12, 215–229. [Google Scholar]
- Lujambio, A.; Portela, A.; Liz, J.; Melo, S.A.; Rossi, S.; Spizzo, R.; Croce, C.M.; Calin, G.A.; Esteller, M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene 2010, 29, 6390–6401. [Google Scholar]
- Hirotsune, S.; Yoshida, N.; Chen, A.; Garrett, L.; Sugiyama, F.; Takahashi, S.; Yagami, K.; Wynshaw-Boris, A.; Yoshiki, A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature 2003, 423, 91–96. [Google Scholar]
- Hawkins, P.G.; Morris, K.V. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 2010, 1, 165–175. [Google Scholar]
- Harrison, P.M.; Zheng, D.; Zhang, Z.; Carriero, N.; Gerstein, M. Transcribed processed pseudogenes in the human genome: An intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Res 2005, 33, 2374–2383. [Google Scholar]
- Pink, R.C.; Wicks, K.; Caley, D.P.; Punch, E.K.; Jacobs, L.; Carter, D.R. Pseudogenes: Pseudo-functional or key regulators in health and disease? RNA 2011, 17, 792–798. [Google Scholar]
- He, L. Posttranscriptional regulation of PTEN dosage by noncoding RNAs. Sci. Signal 2010, 3, pe39. [Google Scholar]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar]
- Ledford, H. Circular RNAs throw genetics for a loop. Nature 2013, 494, 415. [Google Scholar]
- Kosik, K.S. Molecular biology: Circles reshape the RNA world. Nature 2013, 495, 322–324. [Google Scholar]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar]
- Mancini-Dinardo, D.; Steele, S.J.; Levorse, J.M.; Ingram, R.S.; Tilghman, S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev 2006, 20, 1268–1282. [Google Scholar]
- Pauler, F.M.; Koerner, M.V.; Barlow, D.P. Silencing by imprinted noncoding RNAs: Is transcription the answer? Trends Genet 2007, 23, 284–292. [Google Scholar]
- Engstrom, P.G.; Suzuki, H.; Ninomiya, N.; Akalin, A.; Sessa, L.; Lavorgna, G.; Brozzi, A.; Luzi, L.; Tan, S.L.; Yang, L.; et al. Complex Loci in human and mouse genomes. PLoS Genet 2006, 2, e47. [Google Scholar]
- Li, Y.; Sasaki, H. Genomic imprinting in mammals: Its life cycle, molecular mechanisms and reprogramming. Cell Res 2011, 21, 466–473. [Google Scholar]
- Mohammad, F.; Mondal, T.; Kanduri, C. Epigenetics of imprinted long noncoding RNAs. Epigenetics 2009, 4, 277–286. [Google Scholar]
- Nagano, T.; Mitchell, J.A.; Sanz, L.A.; Pauler, F.M.; Ferguson-Smith, A.C.; Feil, R.; Fraser, P. The Air noncoding RNA epigenetically silences transcription by targeting G9a to chromatin. Science 2008, 322, 1717–1720. [Google Scholar]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar]
- Tsai, M.C.; Manor, O.; Wan, Y.; Mosammaparast, N.; Wang, J.K.; Lan, F.; Shi, Y.; Segal, E.; Chang, H.Y. Long noncoding RNA as modular scaffold of histone modification complexes. Science 2010, 329, 689–693. [Google Scholar]
- Schmitz, K.M.; Mayer, C.; Postepska, A.; Grummt, I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev 2010, 24, 2264–2269. [Google Scholar]
- Buske, F.A.; Mattick, J.S.; Bailey, T.L. Potential in vivo roles of nucleic acid triple-helices. RNA Biol 2011, 8, 427–439. [Google Scholar]
- Aguilera, A.; Garcia-Muse, T. R loops: From transcription byproducts to threats to genome stability. Mol. Cell 2012, 46, 115–124. [Google Scholar]
- Jeon, Y.; Lee, J.T. YY1 tethers Xist RNA to the inactive X nucleation center. Cell 2011, 146, 119–133. [Google Scholar]
- Hung, T.; Chang, H.Y. Long noncoding RNA in genome regulation: Prospects and mechanisms. RNA Biol 2010, 7, 582–585. [Google Scholar]
- Bonasio, R.; Tu, S.; Reinberg, D. Molecular signals of epigenetic states. Science 2010, 330, 612–616. [Google Scholar]
- Sun, Q.; Csorba, T.; Skourti-Stathaki, K.; Proudfoot, N.J.; Dean, C. R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 2013, 340, 619–621. [Google Scholar]
- Bertani, S.; Sauer, S.; Bolotin, E.; Sauer, F. The noncoding RNA Mistral activates Hoxa6 and Hoxa7 expression and stem cell differentiation by recruiting MLL1 to chromatin. Mol. Cell 2011, 43, 1040–1046. [Google Scholar]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Solda, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008, 18, 1433–1445. [Google Scholar]
- Pandey, R.R.; Mondal, T.; Mohammad, F.; Enroth, S.; Redrup, L.; Komorowski, J.; Nagano, T.; Mancini-Dinardo, D.; Kanduri, C. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol. Cell 2008, 32, 232–246. [Google Scholar]
- Yao, H.; Brick, K.; Evrard, Y.; Xiao, T.; Camerini-Otero, R.D.; Felsenfeld, G. Mediation of CTCF transcriptional insulation by DEAD-box RNA-binding protein p68 and steroid receptor RNA activator SRA. Genes Dev 2010, 24, 2543–2555. [Google Scholar]
- Wang, K.C.; Yang, Y.W.; Liu, B.; Sanyal, A.; Corces-Zimmerman, R.; Chen, Y.; Lajoie, B.R.; Protacio, A.; Flynn, R.A.; Gupta, R.A.; et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 2011, 472, 120–124. [Google Scholar]
- Feng, J.; Bi, C.; Clark, B.S.; Mady, R.; Shah, P.; Kohtz, J.D. The Evf-2 noncoding RNA is transcribed from the Dlx-5/6 ultraconserved region and functions as a Dlx-2 transcriptional coactivator. Genes Dev 2006, 20, 1470–1484. [Google Scholar]
- Lanz, R.B.; McKenna, N.J.; Onate, S.A.; Albrecht, U.; Wong, J.; Tsai, S.Y.; Tsai, M.J.; O’Malley, B.W. A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 1999, 97, 17–27. [Google Scholar]
- Caretti, G.; Schiltz, R.L.; Dilworth, F.J.; di Padova, M.; Zhao, P.; Ogryzko, V.; Fuller-Pace, F.V.; Hoffman, E.P.; Tapscott, S.J.; Sartorelli, V. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell 2006, 11, 547–560. [Google Scholar]
- Watanabe, M.; Yanagisawa, J.; Kitagawa, H.; Takeyama, K.; Ogawa, S.; Arao, Y.; Suzawa, M.; Kobayashi, Y.; Yano, T.; Yoshikawa, H.; et al. A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor alpha coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J 2001, 20, 1341–1352. [Google Scholar]
- Lanz, R.B.; Chua, S.S.; Barron, N.; Soder, B.M.; DeMayo, F.; O’Malley, B.W. Steroid receptor RNA activator stimulates proliferation as well as apoptosis in vivo. Mol. Cell Biol 2003, 23, 7163–7176. [Google Scholar]
- Kino, T.; Hurt, D.E.; Ichijo, T.; Nader, N.; Chrousos, G.P. Noncoding RNA gas5 is a growth arrest- and starvation-associated repressor of the glucocorticoid receptor. Sci. Signal 2010, 3, ra8. [Google Scholar]
- Willingham, A.T.; Orth, A.P.; Batalov, S.; Peters, E.C.; Wen, B.G.; Aza-Blanc, P.; Hogenesch, J.B.; Schultz, P.G. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 2005, 309, 1570–1573. [Google Scholar]
- Hung, T.; Wang, Y.; Lin, M.F.; Koegel, A.K.; Kotake, Y.; Grant, G.D.; Horlings, H.M.; Shah, N.; Umbricht, C.; Wang, P.; et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat. Genet 2011, 43, 621–629. [Google Scholar]
- Martianov, I.; Ramadass, A.; Serra Barros, A.; Chow, N.; Akoulitchev, A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature 2007, 445, 666–670. [Google Scholar]
- Mazo, A.; Hodgson, J.W.; Petruk, S.; Sedkov, Y.; Brock, H.W. Transcriptional interference: An unexpected layer of complexity in gene regulation. J. Cell Sci 2007, 120, 2755–2761. [Google Scholar]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar]
- Mao, Y.S.; Sunwoo, H.; Zhang, B.; Spector, D.L. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol 2011, 13, 95–101. [Google Scholar]
- Bond, C.S.; Fox, A.H. Paraspeckles: Nuclear bodies built on long noncoding RNA. J. Cell Biol 2009, 186, 637–644. [Google Scholar]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar]
- Chen, L.L.; Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res 2009, 19, 347–359. [Google Scholar]
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol 2012, 9, 1076–1087. [Google Scholar]
- Ip, J.Y.; Nakagawa, S. Long non-coding RNAs in nuclear bodies. Dev Growth Differ 2012, 54, 44–54. [Google Scholar]
- Nakagawa, S.; Naganuma, T.; Shioi, G.; Hirose, T. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol 2011, 193, 31–39. [Google Scholar]
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep 2012, 2, 111–123. [Google Scholar]
- Yin, Q.F.; Yang, L.; Zhang, Y.; Xiang, J.F.; Wu, Y.W.; Carmichael, G.G.; Chen, L.L. Long noncoding RNAs with snoRNA ends. Mol. Cell 2012, 48, 219–230. [Google Scholar]
- Carrieri, C.; Cimatti, L.; Biagioli, M.; Beugnet, A.; Zucchelli, S.; Fedele, S.; Pesce, E.; Ferrer, I.; Collavin, L.; Santoro, C.; et al. Long non-coding antisense RNA controls Uchl1 translation through an embedded SINEB2 repeat. Nature 2012, 491, 454–457. [Google Scholar]
- Yoon, J.H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 suppresses target mRNA translation. Mol. Cell 2012, 47, 648–655. [Google Scholar]
- Kretz, M.; Siprashvili, Z.; Chu, C.; Webster, D.E.; Zehnder, A.; Qu, K.; Lee, C.S.; Flockhart, R.J.; Groff, A.F.; Chow, J.; et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature 2013, 493, 231–235. [Google Scholar]
- Gong, C.; Maquat, L.E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3′ UTRs via Alu elements. Nature 2011, 470, 284–288. [Google Scholar]
- Cesana, M.; Cacchiarelli, D.; Legnini, I.; Santini, T.; Sthandier, O.; Chinappi, M.; Tramontano, A.; Bozzoni, I. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 2011, 147, 358–369. [Google Scholar]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar]
- Spitale, R.C.; Tsai, M.C.; Chang, H.Y. RNA templating the epigenome: Long noncoding RNAs as molecular scaffolds. Epigenetics 2011, 6, 539–543. [Google Scholar]
- Zappulla, D.C.; Cech, T.R. RNA as a flexible scaffold for proteins: Yeast telomerase and beyond. Cold Spring Harb. Symp. Quant. Biol 2006, 71, 217–224. [Google Scholar]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.L.; Nutter, R.C.; Chang, H.Y.; Segal, E. Genome-wide measurement of RNA secondary structure in yeast. Nature 2010, 467, 103–107. [Google Scholar]
- Castello, A.; Fischer, B.; Eichelbaum, K.; Horos, R.; Beckmann, B.M.; Strein, C.; Davey, N.E.; Humphreys, D.T.; Preiss, T.; Steinmetz, L.M.; et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012, 149, 1393–1406. [Google Scholar]
- Guttman, M.; Donaghey, J.; Carey, B.W.; Garber, M.; Grenier, J.K.; Munson, G.; Young, G.; Lucas, A.B.; Ach, R.; Bruhn, L.; et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011, 477, 295–300. [Google Scholar] [Green Version]
- Sheik Mohamed, J.; Gaughwin, P.M.; Lim, B.; Robson, P.; Lipovich, L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 2010, 16, 324–337. [Google Scholar]
- Taft, R.J.; Pang, K.C.; Mercer, T.R.; Dinger, M.; Mattick, J.S. Non-coding RNAs: Regulators of disease. J. Pathol 2010, 220, 126–139. [Google Scholar]
- Ishii, N.; Ozaki, K.; Sato, H.; Mizuno, H.; Saito, S.; Takahashi, A.; Miyamoto, Y.; Ikegawa, S.; Kamatani, N.; Hori, M.; et al. Identification of a novel non-coding RNA, MIAT, that confers risk of myocardial infarction. J. Hum. Genet 2006, 51, 1087–1099. [Google Scholar]
- Pasmant, E.; Laurendeau, I.; Heron, D.; Vidaud, M.; Vidaud, D.; Bieche, I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: Identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res 2007, 67, 3963–3969. [Google Scholar]
- Sonkoly, E.; Bata-Csorgo, Z.; Pivarcsi, A.; Polyanka, H.; Kenderessy-Szabo, A.; Molnar, G.; Szentpali, K.; Bari, L.; Megyeri, K.; Mandi, Y.; et al. Identification and characterization of a novel, psoriasis susceptibility-related noncoding RNA gene, PRINS. J. Biol. Chem 2005, 280, 24159–24167. [Google Scholar]
- Daughters, R.S.; Tuttle, D.L.; Gao, W.; Ikeda, Y.; Moseley, M.L.; Ebner, T.J.; Swanson, M.S.; Ranum, L.P. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 2009, 5, e1000600. [Google Scholar]
- Khalil, A.M.; Faghihi, M.A.; Modarresi, F.; Brothers, S.P.; Wahlestedt, C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLoS One 2008, 3, e1486. [Google Scholar]
- Kogo, R.; Shimamura, T.; Mimori, K.; Kawahara, K.; Imoto, S.; Sudo, T.; Tanaka, F.; Shibata, K.; Suzuki, A.; Komune, S.; et al. Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin modification and is associated with poor prognosis in colorectal cancers. Cancer Res 2011, 71, 6320–6326. [Google Scholar]
- Chung, S.; Nakagawa, H.; Uemura, M.; Piao, L.; Ashikawa, K.; Hosono, N.; Takata, R.; Akamatsu, S.; Kawaguchi, T.; Morizono, T.; et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci 2011, 102, 245–252. [Google Scholar]
- Yang, Z.; Zhou, L.; Wu, L.M.; Lai, M.C.; Xie, H.Y.; Zhang, F.; Zheng, S.S. Overexpression of long non-coding RNA HOTAIR predicts tumor recurrence in hepatocellular carcinoma patients following liver transplantation. Ann. Surg. Oncol 2011, 18, 1243–1250. [Google Scholar]
- Lai, M.C.; Yang, Z.; Zhou, L.; Zhu, Q.Q.; Xie, H.Y.; Zhang, F.; Wu, L.M.; Chen, L.M.; Zheng, S.S. Long non-coding RNA MALAT-1 overexpression predicts tumor recurrence of hepatocellular carcinoma after liver transplantation. Med. Oncol 2012, 29, 1810–1816. [Google Scholar]
- Calin, G.A.; Pekarsky, Y.; Croce, C.M. The role of microRNA and other non-coding RNA in the pathogenesis of chronic lymphocytic leukemia. Best. Pract. Res. Clin. Haematol 2007, 20, 425–437. [Google Scholar]
- Khaitan, D.; Dinger, M.E.; Mazar, J.; Crawford, J.; Smith, M.A.; Mattick, J.S.; Perera, R.J. The melanoma-upregulated long noncoding RNA SPRY4-IT1 modulates apoptosis and invasion. Cancer Res 2011, 71, 3852–3862. [Google Scholar]
- Huarte, M.; Rinn, J.L. Large non-coding RNAs: Missing links in cancer? Hum. Mol. Genet 2010, 19, R152–R161. [Google Scholar]
- Li, L.; Feng, T.; Lian, Y.; Zhang, G.; Garen, A.; Song, X. Role of human noncoding RNAs in the control of tumorigenesis. Proc. Natl. Acad. Sci. USA 2009, 106, 12956–12961. [Google Scholar]
- Yu, W.; Gius, D.; Onyango, P.; Muldoon-Jacobs, K.; Karp, J.; Feinberg, A.P.; Cui, H. Epigenetic silencing of tumour suppressor gene p15 by its antisense RNA. Nature 2008, 451, 202–206. [Google Scholar]
- Lawrie, C.H. MicroRNAs and lymphomagenesis: A functional review. Br. J. Haematol 2013, 160, 571–581. [Google Scholar]
- Agirre, X.; Martinez-Climent, J.A.; Odero, M.D.; Prosper, F. Epigenetic regulation of miRNA genes in acute leukemia. Leukemia 2012, 26, 395–403. [Google Scholar]
- Elton, T.S.; Selemon, H.; Elton, S.M.; Parinandi, N.L. Regulation of the MIR155 host gene in physiological and pathological processes. Gene 2012. [Google Scholar] [CrossRef]
- Vargova, K.; Curik, N.; Burda, P.; Basova, P.; Kulvait, V.; Pospisil, V.; Savvulidi, F.; Kokavec, J.; Necas, E.; Berkova, A.; et al. MYB transcriptionally regulates the miR-155 host gene in chronic lymphocytic leukemia. Blood 2011, 117, 3816–3825. [Google Scholar]
- Nielsen, C.B.; Shomron, N.; Sandberg, R.; Hornstein, E.; Kitzman, J.; Burge, C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 2007, 13, 1894–1910. [Google Scholar]
- Ji, M.; Rao, E.; Ramachandrareddy, H.; Shen, Y.; Jiang, C.; Chen, J.; Hu, Y.; Rizzino, A.; Chan, W.C.; Fu, K.; et al. The miR-17–92 microRNA cluster is regulated by multiple mechanisms in B-cell malignancies. Am. J. Pathol 2011, 179, 1645–1656. [Google Scholar]
- Rinaldi, A.; Poretti, G.; Kwee, I.; Zucca, E.; Catapano, C.V.; Tibiletti, M.G.; Bertoni, F. Concomitant MYC and microRNA cluster miR-17–92 (C13orf25) amplification in human mantle cell lymphoma. Leuk Lymphoma 2007, 48, 410–412. [Google Scholar]
- Hayashita, Y.; Osada, H.; Tatematsu, Y.; Yamada, H.; Yanagisawa, K.; Tomida, S.; Yatabe, Y.; Kawahara, K.; Sekido, Y.; Takahashi, T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res 2005, 65, 9628–9632. [Google Scholar]
- Humphreys, K.J.; Cobiac, L.; Le Leu, R.K.; Van der Hoek, M.B.; Michael, M.Z. Histone deacetylase inhibition in colorectal cancer cells reveals competing roles for members of the oncogenic miR-17–92 cluster. Mol. Carcinog 2013, 52, 459–474. [Google Scholar]
- Kunkeaw, N.; Jeon, S.H.; Lee, K.; Johnson, B.H.; Tanasanvimon, S.; Javle, M.; Pairojkul, C.; Chamgramol, Y.; Wongfieng, W.; Gong, B.; et al. Cell death/proliferation roles for nc886, a non-coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene 2012. [Google Scholar] [CrossRef]
- Treppendahl, M.B.; Qiu, X.; Sogaard, A.; Yang, X.; Nandrup-Bus, C.; Hother, C.; Andersen, M.K.; Kjeldsen, L.; Mollgaard, L.; Hellstrom-Lindberg, E.; et al. Allelic methylation levels of the noncoding VTRNA2–1 located on chromosome 5q31.1 predict outcome in AML. Blood 2012, 119, 206–216. [Google Scholar]
- He, L.; Thomson, J.M.; Hemann, M.T.; Hernando-Monge, E.; Mu, D.; Goodson, S.; Powers, S.; Cordon-Cardo, C.; Lowe, S.W.; Hannon, G.J.; et al. A microRNA polycistron as a potential human oncogene. Nature 2005, 435, 828–833. [Google Scholar]
- Nagoshi, H.; Taki, T.; Hanamura, I.; Nitta, M.; Otsuki, T.; Nishida, K.; Okuda, K.; Sakamoto, N.; Kobayashi, S.; Yamamoto-Sugitani, M.; et al. Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer Res 2012, 72, 4954–4962. [Google Scholar]
- Zeidler, R.; Joos, S.; Delecluse, H.J.; Klobeck, G.; Vuillaume, M.; Lenoir, G.M.; Bornkamm, G.W.; Lipp, M. Breakpoints of Burkitt’s lymphoma t(8;22) translocations map within a distance of 300 kb downstream of MYC. Genes Chromosomes Cancer 1994, 9, 282–287. [Google Scholar]
- Enciso-Mora, V.; Broderick, P.; Ma, Y.; Jarrett, R.F.; Hjalgrim, H.; Hemminki, K.; van den Berg, A.; Olver, B.; Lloyd, A.; Dobbins, S.E.; et al. A genome-wide association study of Hodgkin’s lymphoma identifies new susceptibility loci at 2p16.1 (REL), 8q24.21 and 10p14 (GATA3). Nat. Genet 2010, 42, 1126–1130. [Google Scholar]
- Huppi, K.; Volfovsky, N.; Runfola, T.; Jones, T.L.; Mackiewicz, M.; Martin, S.E.; Mushinski, J.F.; Stephens, R.; Caplen, N.J. The identification of microRNAs in a genomically unstable region of human chromosome 8q24. Mol. Cancer Res 2008, 6, 212–221. [Google Scholar]
- Beck-Engeser, G.B.; Lum, A.M.; Huppi, K.; Caplen, N.J.; Wang, B.B.; Wabl, M. Pvt1-encoded microRNAs in oncogenesis. Retrovirology 2008, 5, 4. [Google Scholar]
- Barsotti, A.M.; Beckerman, R.; Laptenko, O.; Huppi, K.; Caplen, N.J.; Prives, C. p53-Dependent induction of PVT1 and miR-1204. J. Biol. Chem 2011, 287, 2509–2519. [Google Scholar]
- Zhang, X.; Gejman, R.; Mahta, A.; Zhong, Y.; Rice, K.A.; Zhou, Y.; Cheunsuchon, P.; Louis, D.N.; Klibanski, A. Maternally expressed gene 3, an imprinted noncoding RNA gene, is associated with meningioma pathogenesis and progression. Cancer Res 2010, 70, 2350–2358. [Google Scholar]
- Zhou, Y.; Zhang, X.; Klibanski, A. MEG3 noncoding RNA: A tumor suppressor. J. Mol. Endocrinol 2012, 48, R45–R53. [Google Scholar]
- Saitou, M.; Sugimoto, J.; Hatakeyama, T.; Russo, G.; Isobe, M. Identification of the TCL6 genes within the breakpoint cluster region on chromosome 14q32 in T-cell leukemia. Oncogene 2000, 19, 2796–2802. [Google Scholar]
- Zhang, X.; Rice, K.; Wang, Y.; Chen, W.; Zhong, Y.; Nakayama, Y.; Zhou, Y.; Klibanski, A. Maternally expressed gene 3 (MEG3) noncoding ribonucleic acid: Isoform structure, expression, and functions. Endocrinology 2010, 151, 939–947. [Google Scholar]
- Zhou, Y.; Zhong, Y.; Wang, Y.; Zhang, X.; Batista, D.L.; Gejman, R.; Ansell, P.J.; Zhao, J.; Weng, C.; Klibanski, A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem 2007, 282, 24731–24742. [Google Scholar]
- Gejman, R.; Batista, D.L.; Zhong, Y.; Zhou, Y.; Zhang, X.; Swearingen, B.; Stratakis, C.A.; Hedley-Whyte, E.T.; Klibanski, A. Selective loss of MEG3 expression and intergenic differentially methylated region hypermethylation in the MEG3/DLK1 locus in human clinically nonfunctioning pituitary adenomas. J. Clin. Endocrinol. Metab 2008, 93, 4119–4125. [Google Scholar]
- Benetatos, L.; Vartholomatos, G.; Hatzimichael, E. MEG3 imprinted gene contribution in tumorigenesis. Int. J. Cancer 2011, 129, 773–779. [Google Scholar]
- Garding, A.; Bhattacharya, N.; Claus, R.; Ruppel, M.; Tschuch, C.; Filarsky, K.; Idler, I.; Zucknick, M.; Caudron-Herger, M.; Oakes, C.; et al. Epigenetic upregulation of lncRNAs at 13q14.3 in leukemia is linked to the in cis downregulation of a gene cluster that targets NF-kB. PLoS Genet 2013, 9, e1003373. [Google Scholar]
- Takeuchi, S.; Hofmann, W.K.; Tsukasaki, K.; Takeuchi, N.; Ikezoe, T.; Matsushita, M.; Uehara, Y.; Phillip Koeffler, H. Loss of H19 imprinting in adult T-cell leukaemia/lymphoma. Br. J. Haematol 2007, 137, 380–381. [Google Scholar]
- Li, J.; Rhodes, J.C.; Askew, D.S. Evolutionary conservation of putative functional domains in the human homolog of the murine His-1 gene. Gene 1997, 184, 169–176. [Google Scholar]
- Dallosso, A.R.; Hancock, A.L.; Malik, S.; Salpekar, A.; King-Underwood, L.; Pritchard-Jones, K.; Peters, J.; Moorwood, K.; Ward, A.; Malik, K.T.; et al. Alternately spliced WT1 antisense transcripts interact with WT1 sense RNA and show epigenetic and splicing defects in cancer. RNA 2007, 13, 2287–2299. [Google Scholar]
- Ellis, B.C.; Molloy, P.L.; Graham, L.D. CRNDE: A long non-coding RNA involved in CanceR, neurobiology, and development. Front. Genet 2012, 3, 270. [Google Scholar]
- Taskinen, M.; Ranki, A.; Pukkala, E.; Jeskanen, L.; Kaitila, I.; Makitie, O. Extended follow-up of the Finnish cartilage-hair hypoplasia cohort confirms high incidence of non-Hodgkin lymphoma and basal cell carcinoma. Am. J. Med. Genet A 2008, 146A, 2370–2375. [Google Scholar]
- Braconi, C.; Valeri, N.; Kogure, T.; Gasparini, P.; Huang, N.; Nuovo, G.J.; Terracciano, L.; Croce, C.M.; Patel, T. Expression and functional role of a transcribed noncoding RNA with an ultraconserved element in hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2011, 108, 786–791. [Google Scholar]
- Yang, R.; Frank, B.; Hemminki, K.; Bartram, C.R.; Wappenschmidt, B.; Sutter, C.; Kiechle, M.; Bugert, P.; Schmutzler, R.K.; Arnold, N.; et al. SNPs in ultraconserved elements and familial breast cancer risk. Carcinogenesis 2008, 29, 351–355. [Google Scholar]
- Zhang, X.; Lian, Z.; Padden, C.; Gerstein, M.B.; Rozowsky, J.; Snyder, M.; Gingeras, T.R.; Kapranov, P.; Weissman, S.M.; Newburger, P.E. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009, 113, 2526–2534. [Google Scholar]
- Wagner, L.A.; Christensen, C.J.; Dunn, D.M.; Spangrude, G.J.; Georgelas, A.; Kelley, L.; Esplin, M.S.; Weiss, R.B.; Gleich, G.J. EGO, a novel, noncoding RNA gene, regulates eosinophil granule protein transcript expression. Blood 2007, 109, 5191–5198. [Google Scholar]
- Ebralidze, A.K.; Guibal, F.C.; Steidl, U.; Zhang, P.; Lee, S.; Bartholdy, B.; Jorda, M.A.; Petkova, V.; Rosenbauer, F.; Huang, G.; et al. PU.1 expression is modulated by the balance of functional sense and antisense RNAs regulated by a shared cis-regulatory element. Genes Dev 2008, 22, 2085–2092. [Google Scholar]
- Hu, W.; Yuan, B.; Flygare, J.; Lodish, H.F. Long noncoding RNA-mediated anti-apoptotic activity in murine erythroid terminal differentiation. Genes Dev 2011, 25, 2573–2578. [Google Scholar]
- Aoki, K.; Harashima, A.; Sano, M.; Yokoi, T.; Nakamura, S.; Kibata, M.; Hirose, T. A thymus-specific noncoding RNA, Thy-ncR1, is a cytoplasmic riboregulator of MFAP4 mRNA in immature T-cell lines. BMC Mol. Biol 2010, 11, 99. [Google Scholar]
- Marasa, B.S.; Srikantan, S.; Martindale, J.L.; Kim, M.M.; Lee, E.K.; Gorospe, M.; Abdelmohsen, K. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging (Albany NY) 2010, 2, 333–343. [Google Scholar]
- Meyer, K.B.; Maia, A.T.; O’Reilly, M.; Ghoussaini, M.; Prathalingam, R.; Porter-Gill, P.; Ambs, S.; Prokunina-Olsson, L.; Carroll, J.; Ponder, B.A. A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet 2011, 7, e1002165. [Google Scholar]
- Guan, Y.; Kuo, W.L.; Stilwell, J.L.; Takano, H.; Lapuk, A.V.; Fridlyand, J.; Mao, J.H.; Yu, M.; Miller, M.A.; Santos, J.L.; et al. Amplification of PVT1 contributes to the pathophysiology of ovarian and breast cancer. Clin. Cancer Res 2007, 13, 5745–5755. [Google Scholar]
- Palumbo, A.P.; Boccadoro, M.; Battaglio, S.; Corradini, P.; Tsichlis, P.N.; Huebner, K.; Pileri, A.; Croce, C.M. Human homologue of Moloney leukemia virus integration-4 locus (MLVI-4), located 20 kilobases 3′ of the myc gene, is rearranged in multiple myelomas. Cancer Res 1990, 50, 6478–6482. [Google Scholar]
- Borg, A.; Baldetorp, B.; Ferno, M.; Olsson, H.; Sigurdsson, H. c-myc amplification is an independent prognostic factor in postmenopausal breast cancer. Int. J. Cancer 1992, 51, 687–691. [Google Scholar]
- You, L.; Chang, D.; Du, H.Z.; Zhao, Y.P. Genome-wide screen identifies PVT1 as a regulator of Gemcitabine sensitivity in human pancreatic cancer cells. Biochem. Biophys. Res. Commun 2011, 407, 1–6. [Google Scholar]
- Carramusa, L.; Contino, F.; Ferro, A.; Minafra, L.; Perconti, G.; Giallongo, A.; Feo, S. The PVT-1 oncogene is a Myc protein target that is overexpressed in transformed cells. J. Cell. Physiol 2007, 213, 511–518. [Google Scholar]
- Gil, J.; Peters, G. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: All for one or one for all. Nat. Rev. Mol. Cell Biol 2006, 7, 667–677. [Google Scholar]
- Popov, N.; Gil, J. Epigenetic regulation of the INK4b-ARF-INK4a locus: In sickness and in health. Epigenetics 2010, 5, 685–690. [Google Scholar]
- Kotake, Y.; Nakagawa, T.; Kitagawa, K.; Suzuki, S.; Liu, N.; Kitagawa, M.; Xiong, Y. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene 2011, 30, 1956–1962. [Google Scholar]
- Pasmant, E.; Sabbagh, A.; Vidaud, M.; Bieche, I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 2011, 25, 444–448. [Google Scholar]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet 2010, 6, e1001233. [Google Scholar]
- Liu, Y.; Sanoff, H.K.; Cho, H.; Burd, C.E.; Torrice, C.; Mohlke, K.L.; Ibrahim, J.G.; Thomas, N.E.; Sharpless, N.E. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One 2009, 4, e5027. [Google Scholar]
- Cunnington, M.S.; Santibanez Koref, M.; Mayosi, B.M.; Burn, J.; Keavney, B. Chromosome 9p21 SNPs associated with multiple disease phenotypes correlate with anril expression. PLoS Genet 2010, 6, e1000899. [Google Scholar]
- Iacobucci, I.; Sazzini, M.; Garagnani, P.; Ferrari, A.; Boattini, A.; Lonetti, A.; Papayannidis, C.; Mantovani, V.; Marasco, E.; Ottaviani, E.; et al. A polymorphism in the chromosome 9p21 ANRIL locus is associated to Philadelphia positive acute lymphoblastic leukemia. Leuk Res 2011, 35, 1052–1059. [Google Scholar]
- Miyoshi, N.; Wagatsuma, H.; Wakana, S.; Shiroishi, T.; Nomura, M.; Aisaka, K.; Kohda, T.; Surani, M.A.; Kaneko-Ishino, T.; Ishino, F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 2000, 5, 211–220. [Google Scholar]
- Zhang, X.; Zhou, Y.; Mehta, K.R.; Danila, D.C.; Scolavino, S.; Johnson, S.R.; Klibanski, A. A pituitary-derived MEG3 isoform functions as a growth suppressor in tumor cells. J. Clin. Endocrinol. Metab 2003, 88, 5119–5126. [Google Scholar]
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. microRNA-29 can regulate expression of the long non-coding RNA gene MEG3 in hepatocellular cancer. Oncogene 2011, 30, 4750–4756. [Google Scholar]
- Hagan, J.P.; O’Neill, B.L.; Stewart, C.L.; Kozlov, S.V.; Croce, C.M. At least ten genes define the imprinted Dlk1-Dio3 cluster on mouse chromosome 12qF1. PLoS One 2009, 4, e4352. [Google Scholar]
- Schuster-Gossler, K.; Bilinski, P.; Sado, T.; Ferguson-Smith, A.; Gossler, A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev. Dyn 1998, 212, 214–228. [Google Scholar]
- Mondal, T.; Rasmussen, M.; Pandey, G.K.; Isaksson, A.; Kanduri, C. Characterization of the RNA content of chromatin. Genome Res 2010, 20, 899–907. [Google Scholar]
- Gordon, F.E.; Nutt, C.L.; Cheunsuchon, P.; Nakayama, Y.; Provencher, K.A.; Rice, K.A.; Zhou, Y.; Zhang, X.; Klibanski, A. Increased expression of angiogenic genes in the brains of mouse meg3-null embryos. Endocrinology 2010, 151, 2443–2452. [Google Scholar]
- Ying, L.; Huang, Y.; Chen, H.; Wang, Y.; Xia, L.; Chen, Y.; Liu, Y.; Qiu, F. Downregulated MEG3 activates autophagy and increases cell proliferation in bladder cancer. Mol. Biosyst 2013, 9, 407–411. [Google Scholar]
- Dal Bo, M.; Rossi, F.M.; Rossi, D.; Deambrogi, C.; Bertoni, F.; Del Giudice, I.; Palumbo, G.; Nanni, M.; Rinaldi, A.; Kwee, I.; et al. 13q14 deletion size and number of deleted cells both influence prognosis in chronic lymphocytic leukemia. Genes Chromosomes Cancer 2011, 50, 633–643. [Google Scholar]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar]
- Klein, U.; Lia, M.; Crespo, M.; Siegel, R.; Shen, Q.; Mo, T.; Ambesi-Impiombato, A.; Califano, A.; Migliazza, A.; Bhagat, G.; et al. The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell 2010, 17, 28–40. [Google Scholar]
- Lerner, M.; Harada, M.; Loven, J.; Castro, J.; Davis, Z.; Oscier, D.; Henriksson, M.; Sangfelt, O.; Grander, D.; Corcoran, M.M. DLEU2, frequently deleted in malignancy, functions as a critical host gene of the cell cycle inhibitory microRNAs miR-15a and miR-16–1. Exp. Cell Res 2009, 315, 2941–2952. [Google Scholar]
- Coccia, E.M.; Cicala, C.; Charlesworth, A.; Ciccarelli, C.; Rossi, G.B.; Philipson, L.; Sorrentino, V. Regulation and expression of a growth arrest-specific gene (gas5) during growth, differentiation, and development. Mol. Cell Biol 1992, 12, 3514–3521. [Google Scholar]
- Mourtada-Maarabouni, M.; Pickard, M.R.; Hedge, V.L.; Farzaneh, F.; Williams, G.T. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009, 28, 195–208. [Google Scholar]
- Williams, G.T.; Mourtada-Maarabouni, M.; Farzaneh, F. A critical role for non-coding RNA GAS5 in growth arrest and rapamycin inhibition in human T-lymphocytes. Biochem. Soc. Trans 2011, 39, 482–486. [Google Scholar]
- Mourtada-Maarabouni, M.; Hasan, A.M.; Farzaneh, F.; Williams, G.T. Inhibition of human T-cell proliferation by mammalian target of rapamycin (mTOR) antagonists requires noncoding RNA growth-arrest-specific transcript 5 (GAS5). Mol. Pharmacol 2010, 78, 19–28. [Google Scholar]
- Barsyte-Lovejoy, D.; Lau, S.K.; Boutros, P.C.; Khosravi, F.; Jurisica, I.; Andrulis, I.L.; Tsao, M.S.; Penn, L.Z. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res 2006, 66, 5330–5337. [Google Scholar]
- Hibi, K.; Nakamura, H.; Hirai, A.; Fujikake, Y.; Kasai, Y.; Akiyama, S.; Ito, K.; Takagi, H. Loss of H19 imprinting in esophageal cancer. Cancer Res 1996, 56, 480–482. [Google Scholar]
- Fellig, Y.; Ariel, I.; Ohana, P.; Schachter, P.; Sinelnikov, I.; Birman, T.; Ayesh, S.; Schneider, T.; de Groot, N.; Czerniak, A.; et al. H19 expression in hepatic metastases from a range of human carcinomas. J. Clin. Pathol 2005, 58, 1064–1068. [Google Scholar]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS One 2007, 2, e845. [Google Scholar]
- Berteaux, N.; Lottin, S.; Monte, D.; Pinte, S.; Quatannens, B.; Coll, J.; Hondermarck, H.; Curgy, J.J.; Dugimont, T.; Adriaenssens, E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J. Biol. Chem 2005, 280, 29625–29636. [Google Scholar]
- Nunez, C.; Bashein, A.M.; Brunet, C.L.; Hoyland, J.A.; Freemont, A.J.; Buckle, A.M.; Murphy, C.; Cross, M.A.; Lucas, G.; Bostock, V.J.; et al. Expression of the imprinted tumour-suppressor gene H19 is tightly regulated during normal haematopoiesis and is reduced in haematopoietic precursors of patients with the myeloproliferative disease polycythaemia vera. J. Pathol 2000, 190, 61–68. [Google Scholar]
- Bock, O.; Schlue, J.; Kreipe, H. Reduced expression of H19 in bone marrow cells from chronic myeloproliferative disorders. Leukemia 2003, 17, 815–816. [Google Scholar]
- Tessema, M.; Langer, F.; Bock, O.; Seltsam, A.; Metzig, K.; Hasemeier, B.; Kreipe, H.; Lehmann, U. Down-regulation of the IGF-2/H19 locus during normal and malignant hematopoiesis is independent of the imprinting pattern. Int. J. Oncol 2005, 26, 499–507. [Google Scholar]
- Dugimont, T.; Montpellier, C.; Adriaenssens, E.; Lottin, S.; Dumont, L.; Iotsova, V.; Lagrou, C.; Stehelin, D.; Coll, J.; Curgy, J.J. The H19 TATA-less promoter is efficiently repressed by wild-type tumor suppressor gene product p53. Oncogene 1998, 16, 2395–2401. [Google Scholar]
- Farnebo, M.; Bykov, V.J.; Wiman, K.G. The p53 tumor suppressor: A master regulator of diverse cellular processes and therapeutic target in cancer. Biochem. Biophys. Res. Commun 2010, 396, 85–89. [Google Scholar]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar]
- Yoshimizu, T.; Miroglio, A.; Ripoche, M.A.; Gabory, A.; Vernucci, M.; Riccio, A.; Colnot, S.; Godard, C.; Terris, B.; Jammes, H.; et al. The H19 locus acts in vivo as a tumor suppressor. Proc. Natl. Acad. Sci. USA 2008, 105, 12417–12422. [Google Scholar]
- Leighton, P.A.; Saam, J.R.; Ingram, R.S.; Stewart, C.L.; Tilghman, S.M. An enhancer deletion affects both H19 and Igf2 expression. Genes Dev 1995, 9, 2079–2089. [Google Scholar]
- Scaruffi, P.; Stigliani, S.; Moretti, S.; Coco, S.; De Vecchi, C.; Valdora, F.; Garaventa, A.; Bonassi, S.; Tonini, G.P. Transcribed-Ultra Conserved Region expression is associated with outcome in high-risk neuroblastoma. BMC Cancer 2009, 9, 441. [Google Scholar]
- Mestdagh, P.; Fredlund, E.; Pattyn, F.; Rihani, A.; Van Maerken, T.; Vermeulen, J.; Kumps, C.; Menten, B.; de Preter, K.; Schramm, A.; et al. An integrative genomics screen uncovers ncRNA T-UCR functions in neuroblastoma tumours. Oncogene 2010, 29, 3583–3592. [Google Scholar]
- Sana, J.; Hankeova, S.; Svoboda, M.; Kiss, I.; Vyzula, R.; Slaby, O. Expression levels of transcribed ultraconserved regions uc.73 and uc.388 are altered in colorectal cancer. Oncology 2012, 82, 114–118. [Google Scholar]
- Rossi, S.; Sevignani, C.; Nnadi, S.C.; Siracusa, L.D.; Calin, G.A. Cancer-associated genomic regions (CAGRs) and noncoding RNAs: Bioinformatics and therapeutic implications. Mamm. Genome 2008, 19, 526–540. [Google Scholar]
- Ng, D.; Toure, O.; Wei, M.H.; Arthur, D.C.; Abbasi, F.; Fontaine, L.; Marti, G.E.; Fraumeni, J.F., Jr; Goldin, L.R.; Caporaso, N.; et al. Identification of a novel chromosome region, 13q21.33-q22.2, for susceptibility genes in familial chronic lymphocytic leukemia. Blood 2007, 109, 916–925. [Google Scholar]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med 2005, 353, 1793–1801. [Google Scholar]
- Notari, M.; Neviani, P.; Santhanam, R.; Blaser, B.W.; Chang, J.S.; Galietta, A.; Willis, A.E.; Roy, D.C.; Caligiuri, M.A.; Marcucci, G.; et al. A MAPK/HNRPK pathway controls BCR/ABL oncogenic potential by regulating MYC mRNA translation. Blood 2006, 107, 2507–2516. [Google Scholar]
- Du, Q.; Wang, L.; Zhu, H.; Zhang, S.; Xu, L.; Zheng, W.; Liu, X. The role of heterogeneous nuclear ribonucleoprotein K in the progression of chronic myeloid leukemia. Med. Oncol 2010, 27, 673–679. [Google Scholar]
- Tsujimoto, Y.; Gorham, J.; Cossman, J.; Jaffe, E.; Croce, C.M. The t(14;18) chromosome translocations involved in B-cell neoplasms result from mistakes in VDJ joining. Science 1985, 229, 1390–1393. [Google Scholar]
- Ridanpaa, M.; van Eenennaam, H.; Pelin, K.; Chadwick, R.; Johnson, C.; Yuan, B.; vanVenrooij, W.; Pruijn, G.; Salmela, R.; Rockas, S.; et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 2001, 104, 195–203. [Google Scholar]
- Tanaka, R.; Satoh, H.; Moriyama, M.; Satoh, K.; Morishita, Y.; Yoshida, S.; Watanabe, T.; Nakamura, Y.; Mori, S. Intronic U50 small-nucleolar-RNA (snoRNA) host gene of no protein-coding potential is mapped at the chromosome breakpoint t(3;6)(q27;q15) of human B-cell lymphoma. Genes Cells 2000, 5, 277–287. [Google Scholar]
- Zhao, H.; Zhang, X.; Frazao, J.B.; Condino-Neto, A.; Newburger, P.E. HOX antisense lincRNA HOXA-AS2 is an apoptosis repressor in all trans retinoic acid treated NB4 promyelocytic leukemia cells. J. Cell Biochem 2013. [Google Scholar] [CrossRef]
- Paralkar, V.R.; Weiss, M.J. A new ‘Linc’ between noncoding RNAs and blood development. Genes Dev 2011, 25, 2555–2558. [Google Scholar]
- Halvorsen, M.; Martin, J.S.; Broadaway, S.; Laederach, A. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 2010, 6, e1001074. [Google Scholar]
- Wojcik, S.E.; Rossi, S.; Shimizu, M.; Nicoloso, M.S.; Cimmino, A.; Alder, H.; Herlea, V.; Rassenti, L.Z.; Rai, K.R.; Kipps, T.J.; et al. Non-codingRNA sequence variations in human chronic lymphocytic leukemia and colorectal cancer. Carcinogenesis 2010, 31, 208–215. [Google Scholar]
- Scott, L.J.; Mohlke, K.L.; Bonnycastle, L.L.; Willer, C.J.; Li, Y.; Duren, W.L.; Erdos, M.R.; Stringham, H.M.; Chines, P.S.; Jackson, A.U.; et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007, 316, 1341–1345. [Google Scholar]
- Broadbent, H.M.; Peden, J.F.; Lorkowski, S.; Goel, A.; Ongen, H.; Green, F.; Clarke, R.; Collins, R.; Franzosi, M.G.; Tognoni, G.; et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum. Mol. Genet 2008, 17, 806–814. [Google Scholar]
- Morrison, L.E.; Jewell, S.S.; Usha, L.; Blondin, B.A.; Rao, R.D.; Tabesh, B.; Kemper, M.; Batus, M.; Coon, J.S. Effects of ERBB2 amplicon size and genomic alterations of chromosomes 1, 3, and 10 on patient response to trastuzumab in metastatic breast cancer. Genes Chromosomes Cancer 2007, 46, 397–405. [Google Scholar]
- Nupponen, N.N.; Carpten, J.D. Prostate cancer susceptibility genes: Many studies, many results, no answers. Cancer Metastasis Rev 2001, 20, 155–164. [Google Scholar]
- Smedley, D.; Sidhar, S.; Birdsall, S.; Bennett, D.; Herlyn, M.; Cooper, C.; Shipley, J. Characterization of chromosome 1 abnormalities in malignant melanomas. Genes Chromosomes Cancer 2000, 28, 121–125. [Google Scholar]
- Kagami, M.; O'Sullivan, M.J.; Green, A.J.; Watabe, Y.; Arisaka, O.; Masawa, N.; Matsuoka, K.; Fukami, M.; Matsubara, K.; Kato, F.; et al. The IG-DMR and the MEG3-DMR at human chromosome 14q32.2: Hierarchical interaction and distinct functional properties as imprinting control centers. PLoS Genet 2010, 6, e1000992. [Google Scholar]
- Astuti, D.; Latif, F.; Wagner, K.; Gentle, D.; Cooper, W.N.; Catchpoole, D.; Grundy, R.; Ferguson-Smith, A.C.; Maher, E.R. Epigenetic alteration at the DLK1-GTL2 imprinted domain in human neoplasia: Analysis of neuroblastoma, phaeochromocytoma and Wilms’ tumour. Br. J. Cancer 2005, 92, 1574–1580. [Google Scholar]
- Benetatos, L.; Hatzimichael, E.; Dasoula, A.; Dranitsaris, G.; Tsiara, S.; Syrrou, M.; Georgiou, I.; Bourantas, K.L. CpG methylation analysis of the MEG3 and SNRPN imprinted genes in acute myeloid leukemia and myelodysplastic syndromes. Leuk Res 2010, 34, 148–153. [Google Scholar]
- Khoury, H.; Suarez-Saiz, F.; Wu, S.; Minden, M.D. An upstream insulator regulates DLK1 imprinting in AML. Blood 2010, 115, 2260–2263. [Google Scholar]
- Rojas, J.J.; Guedan, S.; Searle, P.F.; Martinez-Quintanilla, J.; Gil-Hoyos, R.; Alcayaga-Miranda, F.; Cascallo, M.; Alemany, R. Minimal RB-responsive E1A promoter modification to attain potency, selectivity, and transgene-arming capacity in oncolytic adenoviruses. Mol. Ther 2010, 18, 1960–1971. [Google Scholar]
- Castello, A.; Horos, R.; Strein, C.; Fischer, B.; Eichelbaum, K.; Steinmetz, L.M.; Krijgsveld, J.; Hentze, M.W. System-wide identification of RNA-binding proteins by interactome capture. Nat. Protoc 2013, 8, 491–500. [Google Scholar]
- Fan, M.; Li, X.; Jiang, W.; Huang, Y.; Li, J.; Wang, Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp. Ther. Med 2013, 5, 1143–1146. [Google Scholar]
- Chung, C.C.; Chanock, S.J. Current status of genome-wide association studies in cancer. Hum. Genet 2011, 130, 59–78. [Google Scholar]
- Giacomini, K.M.; Brett, C.M.; Altman, R.B.; Benowitz, N.L.; Dolan, M.E.; Flockhart, D.A.; Johnson, J.A.; Hayes, D.F.; Klein, T.; Krauss, R.M.; et al. The pharmacogenetics research network: From SNP discovery to clinical drug response. Clin. Pharmacol. Ther 2007, 81, 328–345. [Google Scholar]
- Mattick, J.S. The genetic signatures of noncoding RNAs. PLoS Genet 2009, 5, e1000459. [Google Scholar]
- Prensner, J.R.; Iyer, M.K.; Balbin, O.A.; Dhanasekaran, S.M.; Cao, Q.; Brenner, J.C.; Laxman, B.; Asangani, I.A.; Grasso, C.S.; Kominsky, H.D.; et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat. Biotechnol 2011, 29, 742–749. [Google Scholar]
- Matouk, I.J.; Abbasi, I.; Hochberg, A.; Galun, E.; Dweik, H.; Akkawi, M. Highly upregulated in liver cancer noncoding RNA is overexpressed in hepatic colorectal metastasis. Eur. J. Gastroenterol. Hepatol 2009, 21, 688–692. [Google Scholar]
- Panzitt, K.; Tschernatsch, M.M.; Guelly, C.; Moustafa, T.; Stradner, M.; Strohmaier, H.M.; Buck, C.R.; Denk, H.; Schroeder, R.; Trauner, M.; et al. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 2007, 132, 330–342. [Google Scholar]
- De Kok, J.B.; Verhaegh, G.W.; Roelofs, R.W.; Hessels, D.; Kiemeney, L.A.; Aalders, T.W.; Swinkels, D.W.; Schalken, J.A. DD3(PCA3), a very sensitive and specific marker to detect prostate tumors. Cancer Res 2002, 62, 2695–2698. [Google Scholar]
- Ifere, G.O.; Ananaba, G.A. Prostate cancer gene expression marker 1 (PCGEM1): A patented prostate- specific non-coding gene and regulator of prostate cancer progression. Recent Pat. DNA Gene Seq 2009, 3, 151–163. [Google Scholar]
- Lee, G.L.; Dobi, A.; Srivastava, S. Prostate cancer: Diagnostic performance of the PCA3 urine test. Nat. Rev. Urol 2011, 8, 123–124. [Google Scholar]
- Tomlins, S.A.; Aubin, S.M.; Siddiqui, J.; Lonigro, R.J.; Sefton-Miller, L.; Miick, S.; Williamsen, S.; Hodge, P.; Meinke, J.; Blase, A.; et al. Urine TMPRSS2:ERG fusion transcript stratifies prostate cancer risk in men with elevated serum PSA. Sci. Transl. Med 2011, 3, 94r, a72.. [Google Scholar]
- Durand, X.; Moutereau, S.; Xylinas, E.; de la Taille, A. Progensa PCA3 test for prostate cancer. Expert Rev. Mol. Diagn 2011, 11, 137–144. [Google Scholar]
- Niinuma, T.; Suzuki, H.; Nojima, M.; Nosho, K.; Yamamoto, H.; Takamaru, H.; Yamamoto, E.; Maruyama, R.; Nobuoka, T.; Miyazaki, Y.; et al. Upregulation of miR-196a and HOTAIR drive malignant character in gastrointestinal stromal tumors. Cancer Res 2012, 72, 1126–1136. [Google Scholar]
- Geng, Y.J.; Xie, S.L.; Li, Q.; Ma, J.; Wang, G.Y. Large intervening non-coding RNA HOTAIR is associated with hepatocellular carcinoma progression. J. Int. Med. Res 2011, 39, 2119–2128. [Google Scholar]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar]
- Huang, K.C.; Rao, P.H.; Lau, C.C.; Heard, E.; Ng, S.K.; Brown, C.; Mok, S.C.; Berkowitz, R.S.; Ng, S.W. Relationship of XIST expression and responses of ovarian cancer to chemotherapy. Mol. Cancer Ther 2002, 1, 769–776. [Google Scholar]
- Tsai, M.C.; Spitale, R.C.; Chang, H.Y. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res 2011, 71, 3–7. [Google Scholar]


| LncRNAs | LOCATION | HEMATOLOGIC DISEASE/SYSTEM | FUNCTION | MOLECULAR MECHANISM | MECHANISMS INVOLVED IN DYSREGULATION | CITATIONS |
|---|---|---|---|---|---|---|
| MIR155HG BIC | 21q21.3 | Burkitt, Hodgkin lymphoma, AML, CLL | Host of miRNAs | miR-155 | Target MYB and NFKB | [134] |
| MIR17HG | 13q31.3 | B-cell lymphoma, MCL | Host of miRNAs | miR-17-92 | Target MYC | [136,137,142] |
| vtRNA2-1 | 5q31.1 | AML (poor prognosis) | PKR inhibition | DNA methylation Deletion 5q | [140,141] | |
| PVT1 | 8q24.21 | MM, Burkitt Lymphoma, T-cell Leukemia, CLL | Oncogene and host of miRNAs | miR-1204 MYC activation | Translocation t(8;14)(q24;q11) t(2;8)(p11;q24) t(8;22)(q24;q11) | [143–148] |
| CDKN2BAS1/ ANRIL | 9p21.3 | AML, ALL | Oncogene | PRC1 and PRC2 targeting | rs3731217-G SNP Deletion, hypermethylation | [26,128,149–151] |
| MEG3 | 14q32.2 | AML, MM | Tumor suppressor | PRC2 binding to control DLK1 imprinting. p53 activation. | DNA methylation | [149,150,152–155] |
| DLEU1/DLEU2 | 13q14.2 | CLL, MM, Lymphoma | Tumor suppressor | hsa-miR-16-1 and 15a BCL2 targeting. NFKB activation | Histone modification, DNA methylation, deletion | [156] |
| GAS5 | 1q25.1 | B-cell Lymphoma, Leukemia | Tumor suppressor | Glucorticoid receptor repression. Regulated by mTOR pathway. | Translocation (1;3)(q25;q27) | [87] |
| H19 | 11p15.5 | AML, CML, MPN, T-cell Leukemia, Lymphoma | Oncogene/tumor suppressor | Activated by Myc and down-regulated by p53. miR-675 targeting Rb | Loss of imprinting | [157] |
| T-UCRs | CLL (prognosis marker) | Oncogene/tumor suppressor | miR control | [46] | ||
| lincRNA-p21 | Not annotated in human | ALL, CML | Tumor suppressor | Activated by p53 binds hnRNP K to induce apoptosis | Not known | [158] |
| TCL-6 | 14q32.13 | T cell leukemia | Poorly characterized | Not described | Translocation and inversions with TCR | [151] |
| WT1-AS | 11p13 | AML, ALL | Poorly characterized | WT-1 control | Not known | [159] |
| CRNDE | 16q12.2 | AML, MM, T-cell leukemia | Oncogene | PRC2 and COREST binding | Not known | [160] |
| RMRP | 9p13.3 | Non-Hodgkin lymphoma | Poorly characterized | Not described | Mutation | [161] |
| SNHG5 | 6q14.3 | B-cell Lymphoma | Poorly characterized | snoRNA host | Translocation (1;3)(q25;q27) | [162] |
| HOXA-AS2 | 7p15.2 | APL | Poorly characterized | Not known | [163] | |
| HOTAIRM1 | 7p15.2 | Hematopoietic regulator | Regulator of myelopoiesis | HOX A genes. | [164] | |
| EGOT | 3p26.1 | Hematopoietic regulator | Regulator of eosinophil development | [165] | ||
| PU.1-AS | 11p11.2 Non annotated | Hematopoietic regulator | PU.1-AS regulate the hematopoiesis regulator PU.1 | PU.1 control | [166] | |
| EPS | Mouse 4qC7 | Hematopoietic regulator | Regulator of erytropoyesis | Pycard repression | [167] | |
| ThyncR1 | 1q23.1 | Hematopoietic regulator | Regulator of T cell selection and maduration. | Riboregulator | [168] |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Garitano-Trojaola, A.; Agirre, X.; Prósper, F.; Fortes, P. Long Non-Coding RNAs in Haematological Malignancies. Int. J. Mol. Sci. 2013, 14, 15386-15422. https://doi.org/10.3390/ijms140815386
Garitano-Trojaola A, Agirre X, Prósper F, Fortes P. Long Non-Coding RNAs in Haematological Malignancies. International Journal of Molecular Sciences. 2013; 14(8):15386-15422. https://doi.org/10.3390/ijms140815386
Chicago/Turabian StyleGaritano-Trojaola, Andoni, Xabier Agirre, Felipe Prósper, and Puri Fortes. 2013. "Long Non-Coding RNAs in Haematological Malignancies" International Journal of Molecular Sciences 14, no. 8: 15386-15422. https://doi.org/10.3390/ijms140815386
APA StyleGaritano-Trojaola, A., Agirre, X., Prósper, F., & Fortes, P. (2013). Long Non-Coding RNAs in Haematological Malignancies. International Journal of Molecular Sciences, 14(8), 15386-15422. https://doi.org/10.3390/ijms140815386