Genetic Isolation among the Northwestern, Southwestern and Central-Eastern Indian Ocean Populations of the Pronghorn Spiny Lobster Panulirus penicillatus
Abstract
:1. Introduction

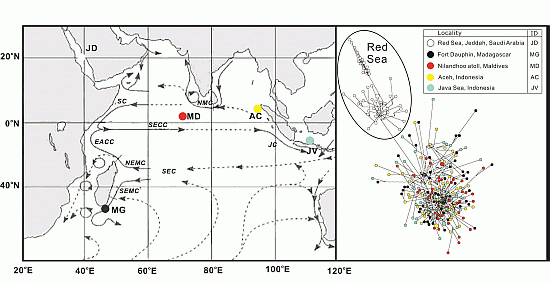
2. Results and Discussion
2.1. Genetic Diversity Analysis
| ID | Locality | N | Year | No. of Haplotypes (Unique) | h ± SD | π ± SD | Tajima’s D (p-Value) | Fu’s F (p-Value) |
|---|---|---|---|---|---|---|---|---|
| JD | Red Sea, Jeddah, Saudi Arabia | 46 | 2011 | 45(45) | 0.9990 ± 0.0048 | 0.031593 ± 0.015851 | −0.79131 (0.215) | −24.20576 (0.000) |
| MG | Fort Dauphin, Madagascar | 49 | 2014 | 49(49) | 1.0000 ± 0.0041 | 0.043441 ± 0.021533 | −0.97966 (0.168) | −24.14011 (0.000) |
| MD | Nilandhoo atoll, Maldives | 38 | 2013 | 37(36) | 0.9986 ± 0.0065 | 0.034957 ± 0.017561 | −1.05781 (0.127) | −19.70407 (0.000) |
| AC | Aceh, Indonesia | 48 | 2011 | 47(47) | 0.9991 ± 0.0045 | 0.038100 ± 0.018974 | −1.17981 (0.098) | −24.14392 (0.000) |
| JV | Java Sea, Indonesia | 55 | 2008 | 53(52) | 0.9993 ± 0.0036 | 0.038855 ± 0.019284 | −1.01996 (0.153) | −24.12306 (0.000) |
| Total | 236 | - | - | - | - | - | - | |
2.2. Population Structure Analysis
| Source of Variation | df | Var. | % Var. | Φ-Statistics | p-Value |
|---|---|---|---|---|---|
| All sites (Red Sea/Madagascar-Maldives-Aceh-Java) | |||||
| Among groups | 1 | 11.50819 | 51.31 | ΦCT = 0.51313 | p = 0.19746 |
| Among populations | 3 | 0.18886 | 0.84 | ΦSC = 0.01730 | p = 0.00098 |
| Within populations | 231 | 10.73030 | 47.84 | ΦST = 0.51313 | p = 0.00000 |
| Substructure grouping test: (Madagascar-Maldives/Aceh/Java) | |||||
| Among groups | 1 | 0.27591 | 2.41 | ΦCT = 0.02408 | p = 0.24047 |
| Among populations | 2 | 0.03850 | 0.34 | ΦSC = 0.00344 | p = 0.14467 |
| Within populations | 186 | 11.14413 | 97.26 | ΦST = 0.02744 | p = 0.00098 |
| (Madagascar/Maldives-Aceh/Java) | |||||
| Among groups | 1 | −0.05540 | −0.49 | ΦCT = −0.00490 | p = 0.67742 |
| Among populations | 2 | 0.21697 | 1.92 | ΦSC = 0.01910 | p = 0.00293 |
| Within populations | 186 | 11.14413 | 98.57 | ΦST = 0.01429 | p = 0.00098 |
| (Madagascar/Aceh-Maldives/Java) | |||||
| Among groups | 1 | −0.08441 | −0.75 | ΦCT = −0.00747 | p = 1.00000 |
| Among populations | 2 | 0.23666 | 2.09 | ΦSC = 0.02079 | p = 0.00098 |
| Within populations | 186 | 11.14413 | 98.65 | ΦST = 0.01348 | p = 0.00098 |
| Population | Northwestern Region (Red Sea) | Southwestern Region (Madagascar) | Central-Eastern Region (Maldives-Aceh-Java) |
|---|---|---|---|
| Northwestern Region (Red Sea) | - | 0.51058 | 0.53463 |
| Southwestern Region (Madagascar) | 0.00000 * | - | 0.02629 |
| Central-Eastern Region (Maldives-Aceh-Java) | 0.00000 * | 0.00098 * | - |

3. Experimental Section
3.1. Lobster Samples
3.2. DNA Analysis
3.3. Genetic Data and Analyses
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Holthuis, L.B. Marine Lobsters of the World: An Annotated and Illustrated Catalogue of Species of Interest to Fisheries Known to Date; Food and Agriculture Organization of the United Nations: Rome, Italy, 1991; Volume 13. [Google Scholar]
- Pitcher, C.R. Spiny lobster. In Nearshore Marine Resources of the South Pacific. Information for Fisheries Development and Management; Wright, A., Hill, L., Eds.; IPS: Suva; Fiji/FFA: Honiara; Solomon/ICOD: Halifax, NS, Canada, 1993; pp. 539–607. [Google Scholar]
- Abdullah, M.F.; Chow, S.; Sakai, M.; Cheng, J.; Imai, H. Genetic diversity and population structure of pronghorn spiny lobster Panulirus penicillatus in the Pacific region. Pac. Sci. 2014, 68, 197–211. [Google Scholar] [CrossRef]
- Booth, J.D.; Phillips, B.F. Early life history of spiny lobster. Crustaceana 1994, 66, 271–294. [Google Scholar] [CrossRef]
- Tolley, K.A.; Groeneveld, J.C.; Gopal, K.; Matthee, C.A. Mitochondrial DNA panmixia in spiny lobster Palinurus gilchristi suggests a population expansion. Mar. Ecol. Prog. Ser. 2005, 297, 225–231. [Google Scholar] [CrossRef]
- Shaklee, J.B.; Bentzen, P. Genetic identification of stocks of marine fish and shellfish. Bull. Mar. Sci. 1998, 62, 589–621. [Google Scholar]
- Diniz, F.M.; Maclean, N.; Ogawa, M.; Cintra, I.H.A.; Bentzen, P. The hypervariable domain of the mitochondrial control region in Atlantic spiny lobsters and its potential as a marker for investigating phylogeographic structuring. Mar. Biotechnol. 2005, 7, 462–473. [Google Scholar] [CrossRef]
- Avise, J.C. Molecular Markers: Natural History and Evolution; Chapman and Hall: London, UK, 1994. [Google Scholar]
- Inoue, N.; Watanabe, H.; Kojima, S.; Sekiguchi, H. Population structure of Japanese spiny lobster Panulirus japonicus inferred by nucleotide sequence analysis of mitochondrial COI gene. Fish. Sci. 2007, 73, 550–556. [Google Scholar] [CrossRef]
- García-Rodríguez, F.J.; Perez-Enriquez, R. Lack of genetic differentiation of blue spiny lobster Panulirus inflatus along the Pacific coast of Mexico inferred from the mtDNA sequences. Mar. Ecol. Prog. Ser. 2008, 361, 203–212. [Google Scholar] [CrossRef]
- Sarver, S.; Silberman, J.D.; Walsh, P.J. Mitochondrial DNA sequence evidence supporting the recognition of two subspecies or species of the Florida spiny lobster Panulirus argus. J. Crust. Biol. 1998, 18, 177–186. [Google Scholar] [CrossRef]
- Farhadi, A.; Farhamand, H.; Nematollahi, M.A.; Jeffs, A.; Lavery, S.D. Mitochondrial DNA population structure of the scalloped lobster Panulirus homarus (Linnaeus 1758) from the West Indian Ocean. ICES J. Mar. Sci. 2013, 70, 1491–1498. [Google Scholar] [CrossRef]
- Chow, S.; Jeff, A.; Miyaeke, Y.; Konishi, K.; Okazaki, M.; Suzuki, N.; Kimura, S.; Abdullah, M.F.; Imai, H.; Sasaki, M.; et al. Genetic isolation between the western and eastern Pacific populations of pronghorn spiny lobster Panulirus penicillatus. PLoS One 2011, 6, e29280. [Google Scholar]
- Schott, F.; McCreary, J.P. The monsoon circulation of the Indian Ocean. Prog. Oceanogr. 2001, 51, 1–123. [Google Scholar] [CrossRef]
- McMillen-Jackson, A.L.; Bert, T.M. Genetic diversity in the mtDNA control region and population structure in the pink shrimp Farfantepenaeus duorarum. J. Crust. Biol. 2004, 24, 101–109. [Google Scholar] [CrossRef]
- McMillen-Jackson, A.L.; Bert, T.M. Disparate patterns of population genetic structure and population history in two sympatric penaeid shrimp species (Farfantepenaeus aztecus and Litopenaeus setiferus) in the eastern United States. Mol. Ecol. 2003, 12, 2895–2905. [Google Scholar] [CrossRef]
- Imai, H.; Hanamura, Y.; Cheng, J.H. Genetic and morphological differentiation in the Sakura shrimp (Sergia lucens) between Japanese and Taiwanese populations. Contrib. Zool. 2013, 82, 123–130. [Google Scholar]
- Heyer, E.; Zietkiewicz, E.; Rochowski, A.; Yotova, V.; Puymirat, J.; Labuda, D. Phylogenetic and familial estimates of mitochondrial substitution rates: study of control region mutations in deep-rooting pedigrees. Am. J. Hum. Genet. 2001, 69, 1113–1126. [Google Scholar] [CrossRef]
- Bird, C.E.; Smouse, P.E.; Karl, S.A.; Toonen, R.J. Detecting and measuring genetic differentiation. In Crustacean Issues: Phylogeography and Population Genetics in Crustacea; Koenemann, S., Held, C., Schubart, C., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 31–55. [Google Scholar]
- Muths, D.; Tessier, E.; Gouws, G.; Craig, M.; Mwale, M.; Mwaluma, J.; Mwandya, A.; Bourjea, J. Restricted dispersal of the reef fish Myripristis berndti at the scale of the SW Indian Ocean. Mar. Ecol. Prog. Ser. 2011, 443, 167–180. [Google Scholar] [CrossRef]
- Castelin, M.; Feutry, P.; Hautecoeur, M.; Marquet, G.; Wowor, D.; Zimmermann, G.; Keith, P. New insight on population genetic connectivity of widespread amphidromous prawn Macrobrachium lar (Fabricius, 1798) (Crustacea: Decapoda: Palaemonidae). Mar. Biol. 2013, 160, 1395–1406. [Google Scholar] [CrossRef]
- Chapman, P.; di Marco, S.F.; Davis, R.E.; Coward, A.C. Flow at intermediate depths around Madagascar based on ALACE float trajectories. Deep Sea Res. Part II Top. Stud. Oceanogr. 2003, 50, 1957–1986. [Google Scholar] [CrossRef]
- Gopurenko, D.; Hughes, J.M.; Keenan, C.P. Mitochondrial DNA evidence for rapid colonisation of the Indo-West Pacific by the mudcrab Scylla serrata. Mar. Biol. 1999, 134, 227–233. [Google Scholar] [CrossRef]
- Fratini, S.; Vannini, M. Genetic differentiation in the mud crab Scylla serrata (Decapoda: Portunidae) within the Indian Ocean. J. Exp. Mar. Biol. Ecol. 2002, 272, 103–116. [Google Scholar] [CrossRef]
- Baums, I.B.; Paris, C.B.; Cherubin, L.M. A biooceanographic filter to larval dispersal in a reef-building coral. Limnol. Oceanogr. 2006, 51, 1969–1981. [Google Scholar] [CrossRef]
- Treml, E.A.; Halpin, P.N.; Urban, D.L.; Pratson, L.F. Modeling population connectivity by ocean currents, a graph-theoretic approach for marine conservation. Landsc. Ecol. 2008, 23, 19–36. [Google Scholar] [CrossRef]
- Riginos, C.; Douglas, K.E.; Jin, Y.; Shanahan, D.F.; Treml, E.A. Effects of geography and life history traits on genetic differentiation in benthic marine fishes. Ecography 2011, 34, 566–575. [Google Scholar] [CrossRef]
- DiBattista, J.D.; Berumen, M.L.; Gaither, M.R.; Rocha, L.A.; Eble, J.A.; Choat, J.H.; Craig, M.T.; Skillings, D.J.; Bowen, B.W. After continents divide: comparative phylogeography of reef fishes from the Red Sea and Indian Ocean. J. Biogeogr. 2013, 40, 1170–1181. [Google Scholar] [CrossRef]
- Johnson, M.W. On the dispersal of lobster larvae into the East Pacific Barrier (Decapoda, Palinuridea). Fish. Bull. 1974, 72, 639–647. [Google Scholar]
- Griffin, D.A.; Wilkin, J.L.; Chubb, C.F.; Pearce, A.F.; Caputi, N. Ocean currents and the larval phase of Australian western rock lobster, Panulirus cygnus. Mar. Freshw. Res. 2001, 52, 1187–1199. [Google Scholar] [CrossRef]
- Butler, M.J., IV; Paris, C.B.; Goldstein, J.S.; Matsuda, H.; Cowen, R.K. Behavior constrains the dispersal of long-lived spiny lobster larvae. Mar. Ecol. Prog. Ser. 2011, 422, 223–237. [Google Scholar] [CrossRef]
- Matsuda, H.; Takenouchi, T.; Goldstein, J.S. The complete larval development of the pronghorn spiny lobster Panulirus penicillatus (Decapoda: Palinuridae) in culture. J. Crust. Biol. 2006, 26, 579–600. [Google Scholar] [CrossRef]
- Shanks, A.L. Pelagic larval duration and dispersal distance revisited. Biol. Bull. 2009, 216, 373–385. [Google Scholar]
- Bradbury, I.R.; Laurel, B.; Snelgrove, P.V.R.; Bentzen, P.; Campana, S.E. Global patterns in marine dispersal estimates: The influence of geography, taxonomic category and life history. Proc. R. Soc. B 2008, 275, 1803–1809. [Google Scholar] [CrossRef]
- Rogers, A.R.; Harpending, H. Population growth waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar]
- Palero, F.; Abelló, P.; Macpherson, E.; Gristina, M.; Pascual, M. Phylogeography of the European spiny lobster (Palinurus elephas): Influence of current oceanographical features and historical processes. Mol. Phylogenet. Evol. 2008, 48, 708–717. [Google Scholar] [CrossRef]
- Babbucci, M.; Buccoli, S.; Cau, A.; Cannas, R.; Goñi, R.; Díaz, D.; Marcato, S.; Zane, L.; Patarnello, T. Population structure, demographic history, and selective processes: Contrasting evidences from mitochondrial and nuclear markers in the European spiny lobster Palinurus elephas (Fabricius, 1787). Mol. Phylogenet. Evol. 2010, 56, 1040–1050. [Google Scholar] [CrossRef]
- Ballard, J.W.; Whitlock, M.C. The incomplete natural history of mitochondria. Mol. Ecol. 2004, 13, 729–744. [Google Scholar] [CrossRef]
- Jiggins, F.M. Male-killing Wolbachia and mitochondrial DNA: Selective sweeps, hybrid introgression and parasite population dynamics. Genetics 2003, 164, 5–12. [Google Scholar]
- Hurst, G.D.D.; Jiggins, F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: The effect of inherited symbionts. Proc. R. Soc. B 2005, 272, 1525–1534. [Google Scholar] [CrossRef]
- Asahida, T.; Kobayashi, T.; Saitoh, K.; Nakayama, I. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish. Sci. 1996, 62, 727–730. [Google Scholar] [CrossRef]
- Imai, H.; Cheng, J.-H.; Hamasaki, K.; Numachi, K.-I. Identification of four mud crab species (genus Scylla) using ITS-1 and 16S rDNA markers. Aquat. Living Resour. 2004, 17, 31–34. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The ClustalX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids. Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Excoffier, L.; Laval, G.; Schneider, S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evol. Bioinform. Online 2005, 1, 47–50. [Google Scholar]
- Rice, W.R. Analyzing tables of statistical tests. Evolution 1989, 43, 223–225. [Google Scholar] [CrossRef]
- Felsenstein, J. PHYLIP-Phylogeny Inference Package. Cladistics 1989, 5, 164–166. [Google Scholar]
- Felsenstein, J. PHYLIP Home Page. Available online: http://evolution.genetics.washington.edu/phylip.html (accessed on 21 May 2014).
- Bandelt, H.J.; Forster, P.; Rohl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Fluxus-engineering.com. Available online: http://www.fluxus-engineering.com (accessed on 21 February 2014).
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar]
- Fu, Y.-X. Statistical tests of neutrality of mutations against population growth, hitchhiking and backgroud selection. Genetics 1997, 147, 915–925. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Abdullah, M.F.; Alimuddin; Muththalib, M.; Salama, A.J.; Imai, H. Genetic Isolation among the Northwestern, Southwestern and Central-Eastern Indian Ocean Populations of the Pronghorn Spiny Lobster Panulirus penicillatus. Int. J. Mol. Sci. 2014, 15, 9242-9254. https://doi.org/10.3390/ijms15069242
Abdullah MF, Alimuddin, Muththalib M, Salama AJ, Imai H. Genetic Isolation among the Northwestern, Southwestern and Central-Eastern Indian Ocean Populations of the Pronghorn Spiny Lobster Panulirus penicillatus. International Journal of Molecular Sciences. 2014; 15(6):9242-9254. https://doi.org/10.3390/ijms15069242
Chicago/Turabian StyleAbdullah, Muhamad Fadry, Alimuddin, Mohamed Muththalib, Adnan Jameel Salama, and Hideyuki Imai. 2014. "Genetic Isolation among the Northwestern, Southwestern and Central-Eastern Indian Ocean Populations of the Pronghorn Spiny Lobster Panulirus penicillatus" International Journal of Molecular Sciences 15, no. 6: 9242-9254. https://doi.org/10.3390/ijms15069242
APA StyleAbdullah, M. F., Alimuddin, Muththalib, M., Salama, A. J., & Imai, H. (2014). Genetic Isolation among the Northwestern, Southwestern and Central-Eastern Indian Ocean Populations of the Pronghorn Spiny Lobster Panulirus penicillatus. International Journal of Molecular Sciences, 15(6), 9242-9254. https://doi.org/10.3390/ijms15069242