Nucleotide-Induced Conformational Changes in Escherichia coli DnaA Protein Are Required for Bacterial ORC to Pre-RC Conversion at the Chromosomal Origin
Abstract
:1. Introduction
2. Results
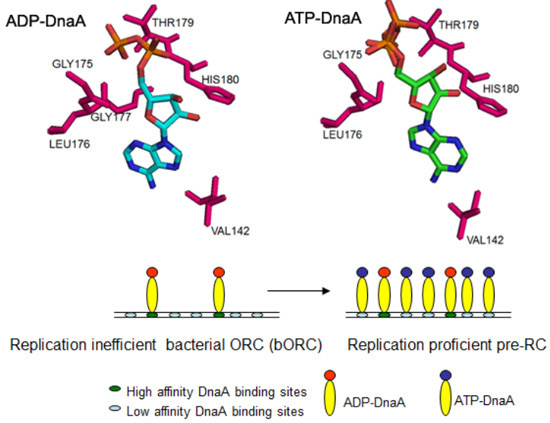
2.1. Conformations of Bound ATP and ADP to DnaA

2.2. ATP-DnaA and ADP-DnaA Have Different Susceptibilities to Proteases


2.3. Proteolytic Susceptibility Sites Predicts Structural Differences between ATP-and ADP-DnaA in Domains II and III
| No. | Peptide Sequence (in Kilo-Daltons) | T1 (45.9) | T3 (32.6) | T4 (30.4) | T5 (19.8) | C2 (29.0) | C3 (18.7) |
|---|---|---|---|---|---|---|---|
| 1 | 13LQDELPATEFSMWIRPLQAELSDNTLALYAPNR45 | - | - | - | X | - | - |
| 2 | 46FVLDWVR52 | X | - | - | X | - | - |
| 3 | 76FEVGTKPVTQTPQAAVTSNVAAPAQVAQTQPQR108 | - | - | - | X | - | X |
| 4 | 115SGWDNVPAPAEPTYR129 | X | - | - | - | X | X |
| 5 | 136HTFDNFVEGK145 | - | X | - | X | X | - |
| 6 | 156QVADNPGGAYNPLFLYGGTGLGKTHLLHAVGNGIMAR192 | X | X | X | X | X | - |
| 7 | 213ALQNNAIEEFKR224 | X | X | X | - | X | - |
| 8 | 228SVDALLIDDIQFFANKERSQEEFFHTFNALLEGNQQIILTSDR273 | X | X | X | - | X | X |
| 9 | 286FGWGLTVAIEPPELETR302 | X | X | X | - | X | X |
| 10 | 309KADENDIRLPGEVAFFIAKR328 | X | X | X | - | X | - |
| 11 | 335ELEGALNRVIANANFTGRAITIDFVR381 | X | X | X | - | X | - |
| 12 | 365DLLALQEKLVTIDNIQK381 | X | X | X | - | X | - |
| 13 | 416ELTNHSLPEIGDAFGGR432 | X | X | X | - | - | - |
| 14 | 448EESHDIKEDFSNLIR463 | X | - | - | - | - | - |

2.4. Adenine Nucleotide Mediated Conformational Changes in DnaA Protein Are Required for Remodeling of Nucleoprotein Complexes at E. coli oriC


3. Discussion
4. Experimental Section
4.1. Reagents and Strains
4.2. Expression and Purification of Recombinant DnaA Protein
4.3. Bioinformatics and Structural Analysis
4.4. Homology Modeling
4.5. Protease Digestion
4.6. Liquid Chromatography Mass Spectroscopy
4.7. DMS Footprinting
4.8. P1 Endonuclease Digestion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fuller, R.S.; Kaguni, J.M.; Kornberg, A. Enzymatic replication of the origin of the Escherichia coli chromosome. Proc. Natl. Acad. Sci. USA 1981, 78, 7370–7374. [Google Scholar] [CrossRef] [PubMed]
- Bramhill, D.; Kornberg, A. Duplex opening by DnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 1988, 52, 743–755. [Google Scholar] [CrossRef]
- Messer, W.; Blaesing, F.; Majka, J.; Nardmann, J.; Schaper, S.; Schmidt, A.; Zakrzewska-Czerwinska, J. Functional domains of DnaA proteins. Biochimie 1999, 81, 819–825. [Google Scholar] [CrossRef]
- Sutton, M.D.; Kaguni, J.M. The Escherichia coli DnaA gene: Four functional domains. J. Mol. Biol. 1997, 274, 546–561. [Google Scholar] [CrossRef] [PubMed]
- Simmons, L.A.; Felczak, M.; Kaguni, J.M. DnaA Protein of Escherichia coli: Oligomerization at the E. coli chromosomal origin is required for initiation and involves specific N-terminal amino acids. Mol. Microbiol. 2003, 49, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Jo, T.; Matsuda, Y.; Matsunaga, C.; Katayama, T.; Ueda, T. Structure and function of DnaA N-terminal domains: Specific sites and mechanisms in inter-DnaA interaction and in DnaB helicase loading on oriC. J. Biol. Chem. 2007, 282, 17816–17827. [Google Scholar] [CrossRef] [PubMed]
- Keyamura, K.; Fujikawa, N.; Ishida, T.; Ozaki, S.; Su’etsugu, M.; Fujimitsu, K.; Katayama, T. The interaction of DiaA and DnaA regulates the replication cycle in E. coli by directly promoting ATP-DnaA-specific initiation complexes. Genes Dev. 2007, 21, 2083–2099. [Google Scholar] [CrossRef] [PubMed]
- Sutton, M.D.; Carr, K.M.; Vicente, M.; Kaguni, J.M. Escherichia coli DnaA protein. The N-terminal domain and loading of DnaB helicase at the E. coli chromosomal origin. J. Biol. Chem. 1998, 273, 34255–34262. [Google Scholar] [CrossRef] [PubMed]
- Seitz, H.; Weigel, C.; Messer, W. The interaction domains of the DnaA and DnaB replication proteins of Escherichia coli. Mol. Microbiol. 2000, 37, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Felczak, M.M.; Simmonst, L.A.; Kaguni, J.M. An essential tryptophan of Escherichia coli DnaA protein functions in oligomerization at the E. coli replication origin. J. Biol. Chem. 2005, 280, 24627–24633. [Google Scholar] [CrossRef] [PubMed]
- Nozaki, S.; Ogawa, T. Determination of the minimum domain II size of Escherichia coli DnaA protein essential for cell viability. Microbiology 2008, 154, 3379–3384. [Google Scholar] [CrossRef] [PubMed]
- Molt, K.L.; Sutera, V.A.; Moore, K.K.; Lovett, S.T. A role for nonessential domain II of initiator protein, DnaA, in replication control. Genetics 2009, 183, 39–49. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.E.; Berger, J.M. AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 2008, 43, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Neuwald, A.F.; Aravind, L.; Spouge, J.L.; Koonin, E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999, 9, 27–43. [Google Scholar] [PubMed]
- Erzberger, J.P.; Pirruccello, M.M.; Berger, J.M. The structure of bacterial DnaA: Implications for general mechanisms underlying DNA replication initiation. EMBO J. 2002, 21, 4763–4773. [Google Scholar] [CrossRef] [PubMed]
- Erzberger, J.P.; Mott, M.L.; Berger, J.M. Structural basis for ATP-dependent DnaA assembly and replication-origin remodeling. Nat. Struct. Mol. Biol. 2006, 13, 676–683. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, T.; Takaki, T.; Kubota, T.; Tsuchiya, T.; Miki, T.; Katayama, T.; Sekimizu, K. Site-directed mutational analysis for the ATP binding of DnaA protein. Functions of two conserved amino acids (Lys-178 and Asp-235) located in the ATP-binding domain of DnaA protein in vitro and in vivo. J. Biol. Chem. 1998, 273, 20847–20851. [Google Scholar] [CrossRef] [PubMed]
- Carr, K.M.; Kaguni, J.M. The A184V missense mutation of the DnaA5 and DnaA46 alleles confers a defect in ATP binding and thermolability in initiation of Escherichia coli DNA replication. Mol. Microbiol. 1996, 20, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Simmons, L.A.; Kaguni, J.M. The DnaAcos allele of Escherichia coli: Hyperactive initiation is caused by substitution of A184V and Y271H, resulting in defective ATP binding and aberrant DNA replication control. Mol. Microbiol. 2003, 47, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, H.; Ozaki, S.; Suzuki, S.; Nakamura, K.; Senriuchi, T.; Su’etsugu, M.; Katayama, T. The exceptionally tight affinity of DnaA for ATP/ADP requires a unique aspartic acid residue in the AAA+ sensor 1 motif. Mol. Microbiol. 2006, 62, 1310–1324. [Google Scholar] [CrossRef] [PubMed]
- Roth, A.; Messer, W. The DNA binding domain of the initiator protein DnaA. EMBO J. 1995, 14, 2106–2111. [Google Scholar] [PubMed]
- Fujikawa, N.; Kurumizaka, H.; Nureki, O.; Terada, T.; Shirouzu, M.; Katayama, T.; Yokoyama, S. Structural basis of replication origin recognition by the DnaA protein. Nucleic Acid Res. 2003, 31, 2077–2086. [Google Scholar] [CrossRef] [PubMed]
- Garner, J.; Crooke, E. Membrane regulation of the chromosomal replication activity of E. coli DnaA requires a discrete site on the protein. EMBO J. 1996, 15, 3477–3485. [Google Scholar] [PubMed]
- Crooke, E. Escherichia coli DnaA protein-phospholipid interactions: In vitro and in vivo. Biochimie 2001, 83, 19–23. [Google Scholar] [CrossRef]
- Matsui, M.; Oka, A.; Takanami, M.; Yasuda, S.; Hirota, Y. Sites of DnaA protein-binding in the replication origin of the Escherichia coli K-12 chromosome. J. Mol. Biol. 1985, 184, 529–533. [Google Scholar] [CrossRef]
- McGarry, K.C.; Ryan, V.T.; Grimwade, J.E.; Leonard, A.C. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc. Natl. Acad. Sci. USA 2004, 10, 2811–2816. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, H.; Keyamura, K.; Katayama, T. Formation of an ATP-DnaA-specific initiation complex requires DnaA Arginine 285, a conserved motif in the AAA+ protein family. J. Biol. Chem. 2005, 280, 27420–27430. [Google Scholar] [CrossRef] [PubMed]
- Grimwade, J.E.; Torgue, J.J.C.; McGarry, K.C.; Rozgaja, T.; Enloe, S.T.; Leonard, A.C. Mutational analysis reveals Escherichia coli oriC interacts with both DnaA-ATP and DnaA-ADP during pre-RC assembly. Mol. Microbiol. 2007, 66, 428–439. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Rozgaja, T.; Grimwade, J.; Crooke, E. Remodeling of nucleoprotein complexes is independent of the nucleotide state of a mutant AAA+ protein. J. Biol. Chem. 2011, 286, 33770–33777. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R.; Fingland, N.; Patil, D.; Sharma, A.; Crooke, E. Crosstalk between DnaA protein, the initiator of Escherichia coli chromosomal replication, and acidic phospholipids present in bacterial membranes. Int. J. Mol. Sci. 2013, 14, 8517–8537. [Google Scholar] [CrossRef] [PubMed]
- Saxena, R. The initiator protein DnaA, chromosomal origin oriC and their interaction to generate origin recognition complex and pre-replication complex like nucleoprotein structures in Escherichia coli. OA Biochem. 2013, 1, 13. [Google Scholar] [CrossRef]
- Rozgaja, T.A.; Grimwade, J.E.; Iqbal, M.; Czerwonka, C.; Vora, M.; Leonard, A.C. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol. Microbiol. 2011, 82, 475–488. [Google Scholar] [CrossRef] [PubMed]
- Kaur, G.; Vora, M.P.; Czerwonka, C.A.; Rozgaja, T.A.; Grimwade, J.E.; Leonard, A.C. Building the bacterial orisome: High-affinity DnaA recognition plays a role in setting the conformation of oriC DNA. Mol. Microbiol. 2014, 91, 1148–1163. [Google Scholar] [CrossRef] [PubMed]
- Duderstadt, K.E.; Mott, M.L.; Crisona, N.J.; Chuang, K.; Yang, H.; Berger, J.M. Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator. J. Biol. Chem. 2010, 285, 28229–28239. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, S.; Kawakami, H.; Nakamura, K.; Fujikawa, N.; Kagawa, W.; Park, S.Y.; Yokoyama, S.; Kurumizaka, H.; Katayama, T. A common mechanism for the ATP-DnaA-dependent formation of open complexes at the replication origin. J. Biol. Chem. 2008, 283, 8351–8362. [Google Scholar] [CrossRef] [PubMed]
- Cassler, M.R.; Grimwade, J.E.; Leonard, A.C. Cell cycle-specific changes in nucleoprotein complexes at a chromosomal replication origin. EMBO J. 1995, 14, 5833–5841. [Google Scholar] [PubMed]
- Miller, D.T.; Grimwade, J.E.; Betteridge, T.; Rozgaja, T.; Torgue, J.J.; Leonard, A.C. Bacterial origin recognition complexes direct assembly of higher-order DnaA oligomeric structures. Proc. Natl. Acad. Sci. USA 2009, 106, 18479–18484. [Google Scholar] [CrossRef] [PubMed]
- Hansen, F.G.; Christensen, B.B.; Nielsen, C.B.; Atlung, T. Insights into the quality of DnaA boxes and their cooperativity. J. Mol. Biol. 2006, 355, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Gon, S.; Camara, J.E.; Klungsøyr, H.K.; Crooke, E.; Skarstad, K.; Beckwith, J. A novel regulatory mechanism couples deoxyribonucleotide synthesis and DNA replication in Escherichia coli. EMBO J. 2006, 25, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Braun, R.E.; O’Day, K.; Wright, A. Autoregulation of the DNA replication gene DnaA in E. coli K-12. Cell 1985, 40, 159–169. [Google Scholar] [CrossRef]
- Kurokawa, K.; Nishida, S.; Emoto, A.; Sekimizu, K.; Katayama, T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. EMBO J. 1999, 18, 6642–6652. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Dowhan, W. In vivo evidence for the involvement of anionic phospholipids in initiation of DNA replication in Escherichia coli. Proc. Natl. Acad. Sci. USA 1995, 92, 783–787. [Google Scholar] [CrossRef] [PubMed]
- Fingland, N.; Flåtten, I.; Downey, C.D.; Fossum-Raunehaug, S.; Skarstad, K.; Crooke, E. Depletion of acidic phospholipids influences chromosomal replication in Escherichia coli. Microbiologyopen 2012, 1, 450–466. [Google Scholar] [CrossRef] [PubMed]
- Heacock, P.N.; Dowhan, W. Alteration of the phospholipid composition of Escherichia coli through genetic manipulation. J. Biol. Chem. 1989, 264, 14972–14977. [Google Scholar] [PubMed]
- Heacock, P.N.; Dowhan, W. Construction of a lethal mutation in the synthesis of the major acidic phospholipids of Escherichia coli. J. Biol. Chem. 1987, 262, 13044–13049. [Google Scholar] [PubMed]
- Sekimizu, K.; Kornberg, A. Cardiolipin activation of DnaA protein, the initiation protein of replication in Escherichia coli. J. Biol. Chem. 1988, 263, 7131–7135. [Google Scholar] [PubMed]
- Castuma, C.E.; Crooke, E.; Kornberg, A. Fluid membranes with acidic domains activate DnaA, the initiator protein of replication in Escherichia coli. J. Biol. Chem. 1993, 268, 24665–24668. [Google Scholar] [PubMed]
- Fujimitsu, K.; Senriuchi, T.; Katayama, T. Specific genomic sequences of Escherichia coli promote replicational initiation by directly reactivating ADP-DnaA. Genes Dev. 2009, 23, 1221–1233. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Kubota, T.; Kurokawa, K.; Crooke, E.; Sekimizu, K. The initiator function of DnaA protein is negatively regulated by the sliding clamp of the Escherichia coli chromosomal replicase. Cell 1998, 94, 61–71. [Google Scholar] [CrossRef]
- Kasho, K.; Katayama, T. DnaA binding locus DatA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc. Natl. Acad. Sci. USA 2013, 110, 936–941. [Google Scholar] [CrossRef] [PubMed]
- Sekimizu, K.; Bramhill, D.; Kornberg, A. ATP activates DnaA protein in initiating replication of plasmids bearing the origin of the Escherichia coli chromosome. Cell 1987, 50, 259–265. [Google Scholar] [CrossRef]
- Sekimizu, K.; Bramhill, D.; Kornberg, A. Sequential early stages in the in vitro initiation of replication at the origin of the Escherichia coli chromosome. J. Biol. Chem. 1988, 263, 7124–7130. [Google Scholar] [PubMed]
- Li, Z.; Crooke, E. Functional analysis of affinity-purified polyhistidine-tagged DnaA protein. Protein Expr. Purif. 1999, 17, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Szpikowska, B.K.; Swiderek, K.M.; Sherman, M.A.; Mas, M.T. MgATP binding to the nucleotide-binding domains of the eukaryotic cytoplasmic chaperonin induces conformational changes in the putative substrate-binding domains. Protein Sci. 1998, 7, 1524–1530. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saxena, R.; Vasudevan, S.; Patil, D.; Ashoura, N.; Grimwade, J.E.; Crooke, E. Nucleotide-Induced Conformational Changes in Escherichia coli DnaA Protein Are Required for Bacterial ORC to Pre-RC Conversion at the Chromosomal Origin. Int. J. Mol. Sci. 2015, 16, 27897-27911. https://doi.org/10.3390/ijms161126064
Saxena R, Vasudevan S, Patil D, Ashoura N, Grimwade JE, Crooke E. Nucleotide-Induced Conformational Changes in Escherichia coli DnaA Protein Are Required for Bacterial ORC to Pre-RC Conversion at the Chromosomal Origin. International Journal of Molecular Sciences. 2015; 16(11):27897-27911. https://doi.org/10.3390/ijms161126064
Chicago/Turabian StyleSaxena, Rahul, Sona Vasudevan, Digvijay Patil, Norah Ashoura, Julia E. Grimwade, and Elliott Crooke. 2015. "Nucleotide-Induced Conformational Changes in Escherichia coli DnaA Protein Are Required for Bacterial ORC to Pre-RC Conversion at the Chromosomal Origin" International Journal of Molecular Sciences 16, no. 11: 27897-27911. https://doi.org/10.3390/ijms161126064
APA StyleSaxena, R., Vasudevan, S., Patil, D., Ashoura, N., Grimwade, J. E., & Crooke, E. (2015). Nucleotide-Induced Conformational Changes in Escherichia coli DnaA Protein Are Required for Bacterial ORC to Pre-RC Conversion at the Chromosomal Origin. International Journal of Molecular Sciences, 16(11), 27897-27911. https://doi.org/10.3390/ijms161126064