Engineering of Self-Assembled Fibronectin Matrix Protein and Its Effects on Mesenchymal Stem Cells
Abstract
:1. Introduction
2. Results and Discussion
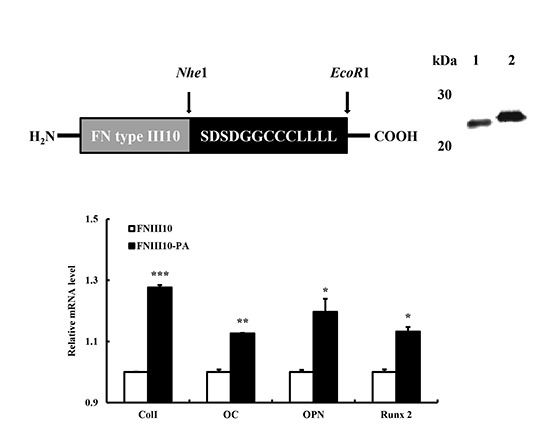
2.1. Expression and Purification of FNIII10-PA in Escherichia coli (E. coli)

2.2. Protein Adhesion Activity of FNIII10-PA

2.3. rMSCs Adhesion Activity Promotion by FNIII10-PA

2.4. The Promotion of rMSCs Proliferation Activity by FNIII10-PA

2.5. Osteogenic Differentiation Gene Eexpression by FNIII10-PA

3. Experimental Section
3.1. Construction of Expression Plasmids
3.2. Protein Expression and Purification
3.3. FNIII10-PA Morphology
3.4. Protein Adhesion Assay
3.5. Cell Isolation and Culture
3.6. Cell Adhesion Assay
3.7. DAPI and F-Actin Staining
3.8. Cell Proliferation Assay
3.9. Quantitative Real-Time PCR Analysis
| Genes | Forward Primer | Reverse Primer |
|---|---|---|
| GAPDH | 5′-TGGAAGGACTCATGACCACA-3′ | 5′-TTCAGCTCAGGGATGACCTT-3′ |
| Col I | 5′-CTGGCAAGAACGGAGATGAT-3′ | 5′-TTAGGACCAGCAGGACCAGT-3′ |
| OC | 5′-CATCACTGCCACCCAGAAGAC-3′ | 5′-CAGTGGATGCAGGGATGATGT-3′ |
| OPN | 5′-CCAATGAAAGCCATGACCAC-3′ | 5′-CGACTGTAGGGACGATTGGA-3′ |
| Runx2 | 5′-CGCCCCTCCCTGAACTCT-3′ | 5′-TGCCTGCCTGGGATCTGTA-3′ |
3.10. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Hur, K.; Hennig, R.G.; Escobedo, F.A.; Wiesner, U. Predicting chiral nanostructures, lattices and superlattices in complex multicomponent nanoparticle self-assembly. Nano Lett. 2012, 12, 3218–3223. [Google Scholar] [CrossRef] [PubMed]
- Maude, S.; Ingham, E.; Aggeli, A. Biomimetic self-assembling peptides as scaffolds for soft tissue engineering. Nanomedicine 2013, 8, 823–847. [Google Scholar] [CrossRef] [PubMed]
- Colombo, G.; Soto, P.; Gazit, E. Peptide self-assembly at the nanoscale: A challenging target for computational and experimental biotechnology. Trends Biotechnol. 2007, 25, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Dinca, V.; Kasotakis, E.; Catherine, J.; Mourka, A.; Ranella, A.; Ovsianikov, A.; Chichkov, B.N.; Farsari, M.; Mitraki, A.; Fotakis, C. Directed three-dimensional patterning of self-assembled peptide fibrils. Nano Lett. 2007, 8, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Konno, T.; Murata, K.; Nagayama, K. Amyloid-like aggregates of a plant protein: A case of a sweet-tasting protein, monellin. FEBS Lett. 1999, 454, 122–126. [Google Scholar] [CrossRef]
- Jarrett, J.T., Jr.; Lansbury, P.T. Amyloid fibril formation requires a chemically discriminating nucleation event: Studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry 1992, 31, 12345–12352. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S. Building from the bottom up. Mater. Today 2003, 6, 20–27. [Google Scholar] [CrossRef]
- Mujeeb, A.; Miller, A.F.; Saiani, A.; Gough, J.E. Self-assembled octapeptide scaffolds for in vitro chondrocyte culture. Acta Biomater. 2013, 9, 4609–4617. [Google Scholar] [CrossRef] [PubMed]
- Gelain, F.; Bottai, D.; Vescovi, A.; Zhang, S. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. PLoS ONE 2006, 27, e119. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.J.; Mayes, A.M.; Griffith, L.G. Nanoscale clustering of RGD peptides at surfaces using Comb polymers. 1. Synthesis and characterization of Comb thin films. Biomacromolecules 2001, 2, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Irvine, D.J.; Ruzette, A.V.; Mayes, A.M.; Griffith, L.G. Nanoscale clustering of RGD peptides at surfaces using comb polymers. 2. Surface segregation of comb polymers in polylactide. Biomacromolecules 2001, 2, 545–556. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, A.; Ling, Q.D.; Kumar, S.S.; Chang, Y.; Kao, T.; Munusamy, M.A.; Alarfaj, A.A.; Hsu, S.; Umezawa, A. External stimulus-responsive biomaterials designed for the culture and differentiation of ES, iPS, and adult stem cells. Prog. Polym. Sci. 2014, 39, 1585–1613. [Google Scholar] [CrossRef]
- Wierzbicka-Patynowski, I.; Schwarzbauer, J.E. The ins and outs of fibronectin matrix assembly. J. Cell Sci. 2003, 116, 3269–3276. [Google Scholar] [CrossRef] [PubMed]
- Oldberg, A.; Linney, E.; Ruoslahti, E. Molecular cloning and nucleotide sequence of a cDNA clone coding for the cell attachment domain in human fibronectin. J. Biol. Chem. 1993, 258, 10193–10196. [Google Scholar]
- Ruoslahti, E. Fibronectin and its receptors. Annu. Rev. Biochem. 1988, 57, 375–413. [Google Scholar] [CrossRef] [PubMed]
- LeBaron, R.G.; Athanasiou, K.A. Extracellular matrix cell adhesion peptides: Functional applications in orthopedic materials. Tissue Eng. 2000, 6, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Hynes, R.O. Integrins: Bidirectional, allosteric signaling machines. Cell 2002, 110, 673–687. [Google Scholar] [CrossRef]
- Kang, W.; Park, S.; Jang, J.H. Kinetic and functional analysis of the heparin-binding domain of fibronectin. Biotechnol. Lett. 2008, 30, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.I.; Jang, J.H.; Chung, C.P.; Ku, Y. Fibronectin fragment promotes osteoblast associated gene expression and biological activity of human osteoblast-like cell. Biotechnol. Lett. 2003, 25, 2007–2011. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.W.; Kang, W.; Jeon, E.; Jang, J.H. Construction and expression of a recombinant fibronectinIII10 protein for integrin-mediated cell adhesion. Biotechnol. Lett. 2010, 32, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Park, S.O.; Jang, H.J.; Jang, J.H. Importance of the heparin-binding domain of fibronectin for enhancing cell adhesion activity of the recombinant fibronectin. Biotechnol. Lett. 2006, 28, 1409–1413. [Google Scholar] [CrossRef] [PubMed]
- Nuttelman, C.R.; Mortisen, D.J.; Henry, S.M.; Anseth, K.S. Attachment of fibronectin to poly(vinyl alcohol) hydrogels promotes NIH3T3 cell adhesion, proliferation, and migration. J. Biomed. Mater. Res. A 2001, 57, 217–223. [Google Scholar] [CrossRef]
- Pendegrass, C.J.; El-Husseiny, M.; Blunn, G.W. The development of fibronectin-functionalized hydroxyapatite coatings to improve dermal fibroblast attachment in vitro. J. Bone Jt. Surg. Br. 2012, 94, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Martino, M.M.; Mochizuki, M.; Rothenfluh, D.A.; Rempel, S.A.; Hubbell, J.A.; Barker, T.H. Controlling integrin specificity and stem cell differentiation in 2D and 3D environments through regulation of fibronectin domain stability. Biomaterials 2009, 30, 1089–1097. [Google Scholar] [CrossRef] [PubMed]
- Ode, A.; Duda, G.N.; Glaeser, J.D.; Matziolis, G.; Frauenschuh, S.; Perk, C.; Wilson, C.J.; Kasper, G. Toward biomimetic materials in bone regeneration: Functional behavior of mesenchymal stem cells on a broad spectrum of extracellular matrix components. J. Biomed. Mater. Res. A 2010, 95, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Weeks, S.; Kulkarni, A.; Smith, H.; Whittall, C.; Yang, Y.; Middleton, J. The effects of chemokine, adhesion and extracellular matrix molecules on binding of mesenchymal stromal cells to poly(l-lactic acid). Cytotherapy 2012, 14, 1080–1088. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, Y.; Haag, J.; Jungebluth, P.; Lundin, V.; Lim, M.L.; Baiguera, S.; Ajalloueian, F.; del Gaudio, C.; Bianco, A.; Moll, G.; et al. Viability and proliferation of rat MSCs on adhesion protein-modified PET and PU scaffolds. Biomaterials 2012, 33, 8094–8103. [Google Scholar] [CrossRef] [PubMed]
- Gajko-Galicka, A. Mutations in type I collagen genes resulting in osteogenesis imperfecta in humans. Acta. Biochim. Pol. 2002, 49, 433–441. [Google Scholar] [PubMed]
- Bharadwaj, S.; Naidu, A.G.; Betageri, G.V.; Prasadarao, N.V.; Naidu, A.S. Milk ribonuclease-enriched lactoferrin induces positive effects on bone turnover markers in postmenopausal women. Osteoporos. Int. 2009, 20, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Bond, S.R.; Lau, A.; Penuela, S.; Sampaio, A.V.; Underhill, T.M.; Laird, D.W.; Naus, C.C. Pannexin 3 is a novel target for Runx2, expressed by osteoblasts and mature growth plate chondrocytes. Bone Miner. Res. 2011, 26, 2911–2922. [Google Scholar] [CrossRef] [PubMed]
- Viereck, V.; Siggelkow, H.; Tauber, S.; Raddatz, D.; Schutze, N.; Hufner, M. Differential regulation of Cbfa1/Runx2 and osteocalcin gene expression by vitamin-D3, dexamethasone, and local growth factors in primary human osteoblasts. J. Cell. Biochem. 2002, 86, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhou, Y.; Zhu, J.; Dong, S.; Li, C.; Xiang, Q. Effect of a novel recombinant protein of fibronectinIII7–10/cadherin 11 EC1–2 on osteoblastic adhesion and differentiation. Biosci. Biotechnol. Biochem. 2009, 73, 1999–2006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, L.; Zhu, J.; Kuang, H.; Dong, S.; Wang, H.; Zhang, X.; Zhou, Y. In vitro observations of self-assembled ECM-mimetic bioceramic nanoreservoir delivering rFN/CDH to modulate osteogenesis. Biomaterials 2012, 33, 7468–7477. [Google Scholar] [CrossRef] [PubMed]
- Bowerman, C.J.; Nilsson, B.L. Self-assembly of amphipathic β-sheet peptides: Insights and applications. Biopolymers 2012, 98, 169–184. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yun, Y.-R.; Pham, L.B.H.; Yoo, Y.-R.; Lee, S.; Kim, H.-W.; Jang, J.-H. Engineering of Self-Assembled Fibronectin Matrix Protein and Its Effects on Mesenchymal Stem Cells. Int. J. Mol. Sci. 2015, 16, 19645-19656. https://doi.org/10.3390/ijms160819645
Yun Y-R, Pham LBH, Yoo Y-R, Lee S, Kim H-W, Jang J-H. Engineering of Self-Assembled Fibronectin Matrix Protein and Its Effects on Mesenchymal Stem Cells. International Journal of Molecular Sciences. 2015; 16(8):19645-19656. https://doi.org/10.3390/ijms160819645
Chicago/Turabian StyleYun, Ye-Rang, Le B. Hang Pham, Yie-Ri Yoo, Sujin Lee, Hae-Won Kim, and Jun-Hyeog Jang. 2015. "Engineering of Self-Assembled Fibronectin Matrix Protein and Its Effects on Mesenchymal Stem Cells" International Journal of Molecular Sciences 16, no. 8: 19645-19656. https://doi.org/10.3390/ijms160819645
APA StyleYun, Y. -R., Pham, L. B. H., Yoo, Y. -R., Lee, S., Kim, H. -W., & Jang, J. -H. (2015). Engineering of Self-Assembled Fibronectin Matrix Protein and Its Effects on Mesenchymal Stem Cells. International Journal of Molecular Sciences, 16(8), 19645-19656. https://doi.org/10.3390/ijms160819645