Longitudinal Claudin Gene Expression Analyses in Canine Mammary Tissues and Thereof Derived Primary Cultures and Cell Lines
Abstract
:1. Introduction
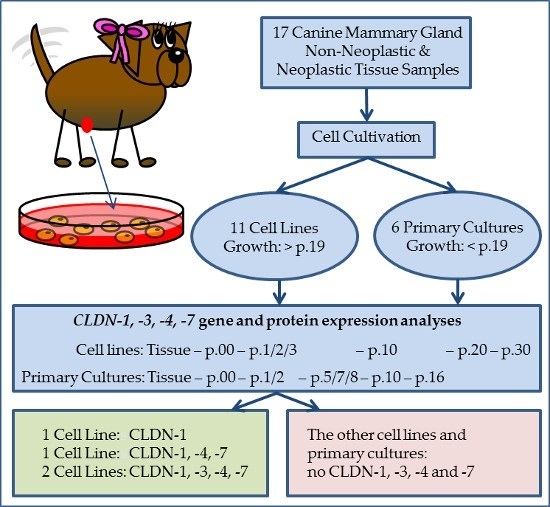
2. Results
2.1. Cell Cultivation and Analyses of the Epithelial Character
2.1.1. Primary Cultures
2.1.2. Cell Lines
2.2. CLDN Gene Expression Analyses
2.2.1. Gene Expression Analyses: QuantiGene Plex Assay
CLDN Gene Expression Alterations from Tissue Samples to Passage 00
CLDN Gene Expressions in the Latest Analysed Passage
CLDN Gene Expression Profiles during Cultivation (Passages 00 until Passage 30)
Linear Correlation of CLDN Expression during Cultivation
2.2.2. Gene Expression Analyses: Conventional PCR
2.3. Immunocytochemistry
2.4. Immunofluorescence
3. Discussion
4. Materials and Methods
4.1. Material, Cell Cultivation and Sample Processing
4.1.1. Canine Mammary Gland Tissue Samples
4.1.2. Establishment of Primary Cultures
4.1.3. Generation of Cell Pellets
4.1.4. Cell Lysis for Gene Expression Analysis Using QuantiGene Plex Assay (Affymetrix)
4.1.5. RNA-Isolation from Tissue Samples for Gene Expression Analyses Using QuantiGene Plex Assay (Affymetrix)
4.1.6. RNA-Isolation from Cultured Cells for Gene Expression Analyses Using Conventional PCR
4.2. Gene Expression Analyses
4.2.1. CLDN Gene Expression Analyses Using QuantiGene Plex Assay (Affymetrix)
CLDN Gene Expression in the Original Tissue Samples and Passages 00
CLDN Expression in the Latest Analysed Passage
Linear Correlation of CLDN Expression during Cultivation
4.2.2. CLDN Gene Expression Analyses Using Conventional PCR
cDNA-Synthesis
CLDN Gene Expression Analyses: Conventional PCR
4.3. Immunocytochemistry
4.4. Immunofluorescence
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Furuse, M.; Fujita, K.; Hiiragi, T.; Fujimoto, K.; Tsukita, S. Claudin-1 and -2: Novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. J. Cell Biol. 1998, 141, 1539–1550. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Sasaki, H.; Fujimoto, K.; Tsukita, S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. J. Cell Biol. 1998, 143, 391–401. [Google Scholar] [CrossRef] [PubMed]
- Günzel, D.; Yu, A.S.L. Claudins and the modulation of tight junction permeability. Physiol. Rev. 2013, 93, 525–569. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, S.L.; Argani, P.; Korz, D.; Evron, E.; Raman, V.; Garrett, E.; Rein, A.; Sauter, G.; Kallioniemi, O.P.; Sukumar, S. Loss of tight junction protein claudin-7 correlates with histological grade in both ductal carcinoma in situ and invasive ductal carcinoma of the breast. Oncogene 2003, 22, 2021–2033. [Google Scholar] [CrossRef] [PubMed]
- Kominsky, S.L.; Vali, M.; Korz, D.; Gabig, T.G.; Weitzman, S.A.; Argani, P.; Sukumar, S. Clostridium perfringens enterotoxin elicits rapid and specific cytolysis of breast carcinoma cells mediated through tight junction proteins claudin 3 and 4. Am. J. Pathol. 2004, 164, 1627–1633. [Google Scholar] [CrossRef]
- Jakab, C.; Halász, J.; Szász, A.M.; Kiss, A.; Schaff, Z.; Rusvai, M.; Gálfi, P.; Kulka, J. Expression of claudin-1, -2, -3, -4, -5 and -7 proteins in benign and malignant canine mammary gland epithelial tumours. J. Comp. Pathol. 2008, 139, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Long, H.; Crean, C.D.; Lee, W.-H.; Cummings, O.W.; Gabig, T.G. Expression of clostridium perfringens enterotoxin receptors claudin-3 and claudin-4 in prostate cancer epithelium. Cancer Res. 2001, 61, 7878–7881. [Google Scholar] [PubMed]
- Krajewska, M.; Olson, A.H.; Mercola, D.; Reed, J.C.; Krajewski, S. Claudin-1 immunohistochemistry for distinguishing malignant from benign epithelial lesions of prostate. Prostate 2007, 67, 907–910. [Google Scholar] [CrossRef] [PubMed]
- Jakab, C.; Rusvai, M.; Demeter, Z.; Gálfi, P.; Szabó, Z.; Kulka, J. Expression of claudin-4 molecule in canine exocrine pancreatic acinar cell carcinomas. Histol. Histopathol. 2011, 26, 1121–1126. [Google Scholar] [PubMed]
- Jakab, C.; Rusvai, M.; Galfi, P.; Halasz, J.; Kulka, J. Expression of claudin-5 in canine pancreatic acinar cell carcinoma—An immunohistochemical study. Acta Vet. Hung. 2011, 59, 87–98. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, S.S.; de Oliveira, I.M.; de Souza, W.; Morgado-Díaz, J.A. Claudins upregulation in human colorectal cancer. FEBS Lett. 2005, 579, 6179–6185. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Semba, S.; Chiba, H.; Sawada, N.; Seo, Y.; Kasuga, M.; Yokozaki, H. Heterogeneous expression of claudin-4 in human colorectal cancer: Decreased claudin-4 expression at the invasive front correlates cancer invasion and metastasis. Pathobiology 2007, 74, 32–41. [Google Scholar] [CrossRef] [PubMed]
- Bornholdt, J.; Friis, S.; Godiksen, S.; Poulsen, S.S.; Santoni-Rugiu, E.; Bisgaardt, H.C.; Lothe, I.M.; Ikdahl, T.; Tveit, K.M.; Johnson, E.; et al. The level of claudin-7 is reduced as an early event in colorectal carcinogenesis. BMC Cancer 2011, 11. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Miyoshi, N.; Ishii, H.; Mimori, K.; Tanaka, F.; Sekimoto, M.; Doki, Y.; Mori, M. Expression of CLDN1 in colorectal cancer: A novel marker for prognosis. Int. J. Oncol. 2011, 39, 791–796. [Google Scholar] [PubMed]
- Jakab, C.; Rusvai, M.; Gálfi, P.; Szabó, Z.; Szabára, Á.; Kulka, J. Expression of claudin-1, -3, -4, -5 and -7 proteins in low grade colorectal carcinoma of canines. Histol. Histopathol. 2010, 25, 55–62. [Google Scholar] [PubMed]
- Sonoda, N.; Furuse, M.; Sasaki, H.; Yonemura, S.; Katahira, J.; Horiguchi, Y.; Tsukita, S. Clostridium perfringens enterotoxin fragment removes specific claudins from tight junction strands: Evidence for direct involvement of claudins in tight junction barrier. J. Cell Biol. 1999, 147, 195–204. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Katahira, J.; Horiguchi, Y.; Sonoda, N.; Furuse, M.; Tsukita, S. Clostridium perfringens enterotoxin binds to the second extracellular loop of claudin-3, a tight junction integral membrane protein. FEBS Lett. 2000, 476, 258–261. [Google Scholar] [CrossRef]
- Santin, A.D.; Cané, S.; Bellone, S.; Palmieri, M.; Siegel, E.R.; Thomas, M.; Roman, J.J.; Burnett, A.; Cannon, M.J.; Pecorelli, S. Treatment of chemotherapy-resistant human ovarian cancer xenografts in C.B-17/SCID mice by intraperitoneal administration of clostridium perfringens enterotoxin. Cancer Res. 2005, 65, 4334–4342. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; McClane, B.A. Use of clostridium perfringens enterotoxin and the enterotoxin receptor-binding domain (C-CPE) for cancer treatment: Opportunities and challenges. J. Toxicol. 2012, 2012, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kondoh, M.; Masuyama, A.; Takahashi, A.; Asano, N.; Mizuguchi, H.; Koizumi, N.; Fujii, M.; Hayakawa, T.; Horiguchi, Y.; Watanbe, Y. A novel strategy for the enhancement of drug absorption using a claudin modulator. Mol. Pharmacol. 2005, 67, 749–756. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, A.; Saito, Y.; Kondoh, M.; Matsushita, K.; Krug, S.M.; Suzuki, H.; Tsujino, H.; Li, X.; Aoyama, H.; Matsuhisa, K.; et al. Creation and biochemical analysis of a broad-specific claudin binder. Biomaterials 2012, 33, 3464–3474. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Lin, X.; Manorek, G.; Kanatani, I.; Cheung, L.H.; Rosenblum, M.G.; Howell, S.B. Recombinant CPE fused to tumor necrosis factor targets human ovarian cancer cells expressing the claudin-3 and claudin-4 receptors. Mol. Cancer Ther. 2009, 8, 1906–1015. [Google Scholar] [CrossRef] [PubMed]
- Mineta, K.; Yamamoto, Y.; Yamazaki, Y.; Tanaka, H.; Tada, Y.; Saito, K.; Tamura, A.; Igarashi, M.; Endo, T.; Takeuchi, K.; et al. Predicted expansion of the claudin multigene family. FEBS Lett. 2011, 585, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Xu, X.; McClane, B.; Zeng, Q.; Litkouhi, B.; Welch, W.R.; Berkowitz, R.S.; Mok, S.C.; Garner, E.I. C-terminus of Clostridium perfringens enterotoxin downregulates CLDN4 and sensitizes ovarian cancer cells to Taxol and Carboplatin. Clin. Cancer Res. 2011, 17, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Michl, P.; Buchholz, M.; Rolke, M.; Kunsch, S.; Löhr, M.; McClane, B.; Tsukita, S.; Leder, G.; Adler, G.; Gress, T.M. Claudin-4: A new target for pancreatic cancer treatment using clostridium perfringens enterotoxin. Gastroenterology 2001, 121, 678–684. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Vicario, F.; Alvarez, C.E.; Rowell, J.L.; Roy, S.; London, C.A.; Kisseberth, W.C.; Lorch, G. Human genetic relevance and potent antitumor activity of heat shock protein 90 inhibition in canine lung adenocarcinoma cell lines. PLoS ONE 2015, 10, e0142007. [Google Scholar] [CrossRef] [PubMed]
- Hammer, S.C.; Nagel, S.; Junginger, J.; Hewicker-Trautwein, M.; Wagner, S.; Heisterkamp, A.; Ngezahayo, A.; Nolte, I.; Murua Escobar, H. Claudin-1, -3, -4 and -7 gene expression analyses in canine prostate carcinoma and mammary tissue derived cell lines. Neoplasma 2016, 63, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Aung, P.P.; Mitani, Y.; Sanada, Y.; Nakayama, H.; Matsusaki, K.; Yasui, W. Differential expression of claudin-2 in normal human tissues and gastrointestinal carcinomas. Virchows Arch. 2006, 448, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Kunisaki, C.; Yoshihara, K.; Yamada, R.; Yamamoto, N.; Sato, T.; Makino, H.; Yamagishi, S.; Nagano, Y.; Fujii, S.; et al. Reduced expression of claudin-7 gene correlates with venous invasion and liver metastasis in colorectal cancer. Oncol. Rep. 2008, 19, 953–959. [Google Scholar] [CrossRef] [PubMed]
- Gruntzig, K.; Graf, R.; Boo, G.; Guscetti, F.; Hassig, M.; Axhausen, K.W.; Fabrikant, S.; Welle, M.; Meier, D.; Folkers, G.; et al. Swiss canine cancer registry 1955–2008: Occurrence of the most common tumour diagnoses and influence of age, breed, body size, sex and neutering status on tumour development. J. Comp. Pathol. 2016, 155, 156–170. [Google Scholar] [CrossRef] [PubMed]
- Sorenmo, K. Canine mammary gland tumours. Vet. Clin. N. Am. Small Anim. Pract. 2003, 33, 573–596. [Google Scholar] [CrossRef]
- Santos, A.A.; Matos, A.J.F. Advances in the understanding of the clinically relevant pathways and molecular aspects of canine mammary tumors. Part 2: Invasion, angiogenesis, metastasis and therapy. Vet. J. 2015, 205, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Abdoon, A.S.; Al-Ashkar, E.A.; Kandil, O.M.; Shaban, A.M.; Khaled, H.M.; El Sayed, M.A.; El Shaer, M.M.; Shaalan, A.H.; Eisa, W.H.; Eldin, A.A.; et al. Efficacy and toxicity of plasmonic photothermal therapy (PPTT) using gold nanorods (GNRs) against mammary tumors in dogs and cats. Nanomedicine 2016, 12, 2291–2297. [Google Scholar] [CrossRef] [PubMed]
- Rowell, J.L.; McCarty, D.O.; Alvarez, C.E. Dog models of naturally occuring cancer. Trends Mol. Med. 2011, 17, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Martin, T.A.; Jiang, W.G. Loss of tight junction barrier function and its role in cancer metastasis. Biochim. Biophys. Acta 2009, 1788, 872–891. [Google Scholar] [CrossRef] [PubMed]
- Escudero-Esparza, A.; Jiang, W.G.; Martin, T.A. The claudin family and its role in cancer and metastasis. Front. Biosci. 2011, 16, 1069–1083. [Google Scholar] [CrossRef]
- Kölbl, A.C.; Jeschke, U.; Andergassen, U. The significance of epithelial-to-mesenchymal transition for circultaing tumor cells. Int. J. Mol. Sci. 2016, 17. [Google Scholar] [CrossRef] [PubMed]
- Furuse, M.; Hiroyuki, S.; Tsukita, S. Manner of interaction of heterogeneous claudin species within and between tight junction strands. J. Cell Biol. 1999, 147, 891–903. [Google Scholar] [CrossRef] [PubMed]
- Jakab, C.; Halasz, J.; Szasz, A.M.; Batmunkh, E.; Kiss, A.; Schaff, Z.; Rusvai, M.; Gálfi, P.; Kulka, J. Expression and localisation of claudin-1, -2, -3, -4, -5, -7 and -10 proteins in the normal canine mammary gland. Acta Vet. Hung. 2008, 56, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Todd, M.C.; Petty, H.M.; King, J.M.; Piana Marshall, B.N.; Sheller, R.A.; Cuevas, M.E. Overexpression and delocalization of claudin-3 protein in MCF-7 and MDA-MB-415 breast cancer cell lines. Oncol. Lett. 2015, 10, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, M.; Pena, L.; Rasotto, R.; Zapulli, V. Classification and grading of canine mammary tumors. Vet. Pathol. 2011, 48, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Borka, K.; Kaliszky, P.; Szabó, E.; Lotz, G.; Kupcsulik, P.; Schaff, Z.; Kiss, A. Claudin expression in pancreatic endocrine tumors as compared with ductal adenocarcinomas. Virchows Arch. 2007, 450, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Junginger, J.; Kummerfeld, M.; Kummrow, M.; Gruetzmacher, K.; Dziallas, P.; Wohlsein, P. Preorbital carcinoma in two kirk’s dik-diks (Madoqua kirkii). J. Comp. Pathol. 2013, 148, 414–418. [Google Scholar] [CrossRef] [PubMed]
- Von Wasielewski, R.; Mengel, M.; Gignac, S.; Wilkens, L.; Werner, M.; Georgii, A. Tyramine amplification technique in routine immunohistochemistry. J. Histochem. Cytochem. 1997, 45, 1455–1459. [Google Scholar] [CrossRef] [PubMed]
- Caceres, S.; Pena, L.; de Andres, P.J.; Illera, M.J.; Lopez, M.S.; Woodward, W.A.; Reuben, J.M.; Illera, J.C. Establishment and characterization of a new cell line of canine inflammatory mammary cancer: IPC-366. PLoS ONE 2015, 10, e0122277. [Google Scholar] [CrossRef] [PubMed]
- Montiani-Ferreira, F.; Kiupel, M.; Muzolon, P.; Truppel, J. Corneal squamous cell carcinoma in a dog: A case report. Vet. Ophtalmol. 2008, 11, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Labelle, P.; Reilly, C.M.; Naydan, D.K.; Labelle, A.L. Immunohistochemical characteristics of normal canine eyes. Vet. Pathol. 2012, 49, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, E.; Gobbi, H.; Saraiva, B.S.; Cassali, G.D. Columnar cell lesions of the canine mammary gland: Pathological features and immunophenotypic analysis. BMC Cancer 2010, 10. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.H.; Lim, H.Y.; Im, K.S.; Kim, J.H.; Sur, J.H. Identification of triple-negative and basal-like canine mammary carcinomas using four basal markers. J. Comp. Pathol. 2013, 148, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Theerawatanasirikul, S.; Suriyaphol, G.; Thanawongnuwech, R.; Sailasuta, A. Histologic morphology and involucrin, filaggrin, and keratin expression in normal canine skin from dogs of different breeds and coat types. J. Vet. Sci. 2012, 13, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Mutterer, J.; Zinck, E. Quick-and-clean article figures with FigureJ. J. Microsc. 2013, 252, 89–91. [Google Scholar] [CrossRef] [PubMed]






| Cell Line | Histological Classif. | Tissue | p.00 | p.01 | p.02 | p.03 | p.05 * | p.07 * | p.08 * | p.10 | p.16 | p.20 | p.30 | p.40 |
| DT14/05R | non-neoplastic mammary gland tissue | X | X | X | X | |||||||||
| DT14/06R | X | X | X | X | X | X | ||||||||
| T124 | lobular hyperplasia | X | X | X | X | |||||||||
| DT14/04T | simple adenoma | X | X | X | X | X | ||||||||
| T121 | complex adenoma | X | X | X | ||||||||||
| T120A | carcinoma, complex type | X | X | X | ||||||||||
| DT14/06T | X | X | X | X | X | X | ||||||||
| DT14/06Ts | X | X | X | X | X | |||||||||
| T126 | carcinoma arising in a benign mixed tumour | X | X | X | X | |||||||||
| DT14/10 | benign mixed mammary tumour | X | X | X | X | |||||||||
| DT15/02T | X | X | X | X | ||||||||||
| Primary Culture | Histological Classif. | Tissue | p.00 | p.01 | p.02 | p.03 * | p.05 | p.07 | p.08 | p.10 | p.16 | p.20 | p.30 | p.40 |
| DT14/04R | non-neoplastic mammary gland tissue | X | X | ** | ** | ** | ||||||||
| DT14/05S2 | X | X | X | ** | ** | ** | ||||||||
| DT14/07R | X | X | X | ** | ** | ** | ||||||||
| DT14/08R | lobular hyperplasia | X | X | X | X | ** | ** | ** | ||||||
| DT14/07T | simple tubular carcinoma | X | X | X | X | ** | ** | ** | ||||||
| DT14/09T | X | X | X | X | ** | ** | ** |
| Primary Culture | Histological Classif. | p.n | AE1/AE3 | MNF116 | HMW | CD5/6 | CK7 | CK10 | CK14 | CK20 |
|---|---|---|---|---|---|---|---|---|---|---|
| DT14/04R | healthy mammary gland tissue | p.08 | − | |||||||
| p.13 | − | |||||||||
| DT14/05S2 | p.04 | − | ||||||||
| later | NA | |||||||||
| DT14/07R | p.03 | − | (+) | − | (+) | − | ||||
| p.07 | + | + | − | + | + | − | + | − | ||
| DT14/08R | lobular hyperplasia | p.02 | (+) | − | ||||||
| p.05 | − | |||||||||
| DT14/07T | simple tubular carcinoma | p.03 | + | + | + | + | (+) | − | + | − |
| p.17 | + | + | + | (+) | − | + | − | |||
| DT14/09T | p.03 | + | (+) | (+) | − | (+) | − | |||
| p.08 | − | |||||||||
| Cell Line | Histological Classif. | p.n | AE1/AE3 | MNF116 | HMW | CK5/6 | CK7 | CK10 | CK14 | CK20 |
|---|---|---|---|---|---|---|---|---|---|---|
| DT14/05R | healthy mammary gland tissue | p.04 | (+) | − | (+) | − | ||||
| p.21 | − | |||||||||
| DT14/06R | p.04 | (+) | (+) | − | (+) | − | ||||
| p.41 | − | NA | NA | NA | NA | NA | NA | NA | ||
| T124 | lobular hyperplasia | p.03 | + | + | + | + | (+) | (+) | + | − |
| p.36 | + | + | + | + | + | − | + | − | ||
| DT14/04T | simple adenoma | p.05 | + | + | (+) | (+) | − | − | − | |
| p.27 | + | + | − | + | + | − | + | − | ||
| T121 | complex adenoma | p.06 | (+) | (+) | (+) | (+) | − | + | − | |
| p.28 | (+) | − | (+) | − | + | − | ||||
| T120A | carcinoma complex type | p.07 | (+) | (+) | (+) | (+) | (+) | − | + | − |
| p.44 | + | + | + | − | + | − | ||||
| DT14/06T | p.04 | (+) | − | (+) | − | |||||
| p.43 | (+) | − | ||||||||
| DT14/06Ts | p.04 | (+) | (+) | (+) | − | (+) | − | |||
| p.33 | (+) | − | ||||||||
| T126 | carcinoma arising in a benign mixed tumour | p.03 | − | (+) | − | |||||
| p.24 | − | |||||||||
| DT14/10 | benign mixed mammary tumour | p.02 | + | + | NA | − | (+) | − | + | − |
| p.31 | + | + | NA | − | + | − | ||||
| DT15/02T | p.04 | + | (+) | + | − | + | − | |||
| p.45 | (+) | − | (+) | − | (+) | − | ||||
| Gene | CLDN-1 | CLDN-3 | CLDN-4 | CLDN-7 | ||||
|---|---|---|---|---|---|---|---|---|
| passage | e.p. | l.p. | e.p. | l.p. | e.p. | l.p. | e.p. | l.p. |
| T120A | + | + | + | + | + | + | + | + |
| T124 | + | + | + | + | + | + | + | + |
| DT14/04T | + | + | + | + | + | + | + | + |
| DT14/04R | + | + | − | + | − | + | − | + |
| DT14/10 | + | + | + | − | − | − | − | − |
| DT14/06T | − | + | − | − | − | − | − | − |
| T121 | − | + | + | − | − | − | + | − |
| Protein | Antibody | Dilution | |
|---|---|---|---|
| ICC | IF | ||
| CLDN-1 | Polyclonal rabbit anti-human CLDN-1 antibody (Thermo Fischer Scientific, Darmstadt, Germany) * | 1:50 | 1:50 |
| CLDN-3 | Polyclonal rabbit anti-mouse CLDN-3 antibody (Thermo Fischer Scientific) * | 1:200 | 1:83 |
| CLDN-4 | Monoclonal mouse anti-human CLDN-4 antibody (Clone 3E2C1, Thermo Fischer Scientific) * | 1:200 | 1:83 |
| CLDN-7 | Polyclonal rabbit anti-human CLDN-7 antibody (Thermo Fischer Scientific) * | 1:200 | 1:125 |
| Pan-CK (CK1-8, CK10, CK13-16, CK19) | Monoclonal mouse anti-human CK (clone AE1/AE3, Dako, Hamburg, Germany) [45,46,47] | 1:500 | - |
| Pan-CK (CK5, CK6, CK8, CK17, CK19) | Monoclonal mouse anti-human CK (clone MNF116, Dako) [46] * | 1:1000 | - |
| Pan-CK (CK1, CK5, CK10, CK14) | Monoclonal mouse anti-human CK, high molecular weight (clone 34bE12, Dako) [48] | 1:500 | - |
| CK5 and CK6 | Monoclonal mouse anti-human CK5/CK6 (clone D5/16B4, Dako) [49] | 1:100 | - |
| CK7 | Monoclonal mouse anti-human CK7 (clone OV-TL12/30, Dako) [47] | 1:100 | - |
| CK10 | Monoclonal mouse anti-human CK10 (clone DE-K10, Dako) [50] * | 1:100 | - |
| CK14 | Polyclonal rabbit anti-human CK14 (Thermo Fischer Scientific) ** | 1:500 | - |
| CK20 | Monoclonal mouse anti-human CK20 (clone Ks20.8, Dako) [47] * | 1:100 | - |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hammer, S.C.; Becker, A.; Rateitschak, K.; Mohr, A.; Lüder Ripoli, F.; Hennecke, S.; Junginger, J.; Hewicker-Trautwein, M.; Brenig, B.; Ngezahayo, A.; et al. Longitudinal Claudin Gene Expression Analyses in Canine Mammary Tissues and Thereof Derived Primary Cultures and Cell Lines. Int. J. Mol. Sci. 2016, 17, 1655. https://doi.org/10.3390/ijms17101655
Hammer SC, Becker A, Rateitschak K, Mohr A, Lüder Ripoli F, Hennecke S, Junginger J, Hewicker-Trautwein M, Brenig B, Ngezahayo A, et al. Longitudinal Claudin Gene Expression Analyses in Canine Mammary Tissues and Thereof Derived Primary Cultures and Cell Lines. International Journal of Molecular Sciences. 2016; 17(10):1655. https://doi.org/10.3390/ijms17101655
Chicago/Turabian StyleHammer, Susanne C., Annegret Becker, Katja Rateitschak, Annika Mohr, Florenza Lüder Ripoli, Silvia Hennecke, Johannes Junginger, Marion Hewicker-Trautwein, Bertram Brenig, Anaclet Ngezahayo, and et al. 2016. "Longitudinal Claudin Gene Expression Analyses in Canine Mammary Tissues and Thereof Derived Primary Cultures and Cell Lines" International Journal of Molecular Sciences 17, no. 10: 1655. https://doi.org/10.3390/ijms17101655
APA StyleHammer, S. C., Becker, A., Rateitschak, K., Mohr, A., Lüder Ripoli, F., Hennecke, S., Junginger, J., Hewicker-Trautwein, M., Brenig, B., Ngezahayo, A., Nolte, I., & Murua Escobar, H. (2016). Longitudinal Claudin Gene Expression Analyses in Canine Mammary Tissues and Thereof Derived Primary Cultures and Cell Lines. International Journal of Molecular Sciences, 17(10), 1655. https://doi.org/10.3390/ijms17101655