UBXD Proteins: A Family of Proteins with Diverse Functions in Cancer
Abstract
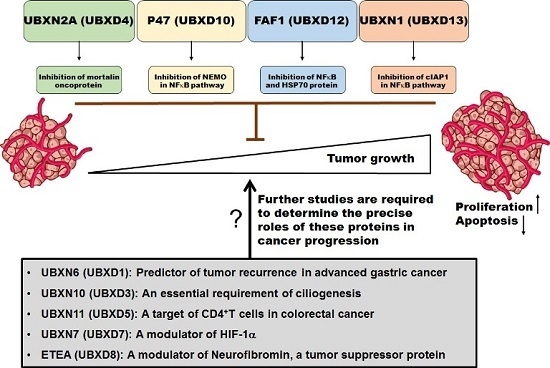
:1. Introduction
2. UBXN6, a Potential Predictor of Postoperative Recurrence in Gastric Cancer
3. UBXN10, Ciliogenesis, and Human Cancer Cells
4. UBXN2A, a Potential Target for Cancer Therapy
5. UBXN11, an Antigen Recognized by Colon Tumor-Reactive T Cells
6. UBXD7, a Selective Modulator of HIF-1α
7. UBXD8, an Inhibitor of Neurofibromin
8. P47, a Negative Regulator of the NF-κB Pathway in Cancer Cells
9. FAF1, a Powerful Anti-Cancer Protein
10. UBXN1, an Inhibitor of Cellular Inhibitors of Apoptosis Proteins (cIAP)
11. Conclusions and Future Directions
Acknowledgments
Conflicts of Interest
References
- Buchberger, A. From UBA to UBX: New words in the ubiquitin vocabulary. Trends Cell Biol. 2002, 12, 216–221. [Google Scholar] [CrossRef]
- Rijal, R.; Arhzaouy, K.; Strucksberg, K.H.; Cross, M.; Hofmann, A.; Schroder, R.; Clemen, C.S.; Eichinger, L. Mutant p97 exhibits species-specific changes of its ATPase activity and compromises the UBXD9-mediated monomerisation of p97 hexamers. Eur. J. Cell Biol. 2016, 95, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Alexandru, G.; Graumann, J.; Smith, G.T.; Kolawa, N.J.; Fang, R.; Deshaies, R.J. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1α turnover. Cell 2008, 134, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Bogan, J.S.; Hendon, N.; McKee, A.E.; Tsao, T.S.; Lodish, H.F. Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 2003, 425, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Orme, C.M.; Bogan, J.S. The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-golgi intermediate compartment. J. Biol. Chem. 2012, 287, 6679–6692. [Google Scholar] [CrossRef] [PubMed]
- Cloutier, P.; Lavallee-Adam, M.; Faubert, D.; Blanchette, M.; Coulombe, B. A newly uncovered group of distantly related lysine methyltransferases preferentially interact with molecular chaperones to regulate their activity. PLoS Genet. 2013, 9, e1003210. [Google Scholar] [CrossRef] [PubMed]
- Alexandru, G. Exploring the role of p97 and its UBX-domain cofactors through identification of their interacting proteins. Methods Mol. Biol. 2012, 832, 305–312. [Google Scholar] [PubMed]
- Bandau, S.; Knebel, A.; Gage, Z.O.; Wood, N.T.; Alexandru, G. UBXN7 docks on neddylated cullin complexes using its UIM motif and causes HIF1α accumulation. BMC Biol. 2012, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Castorena, C.M.; Mackrell, J.G.; Bogan, J.S.; Kanzaki, M.; Cartee, G.D. Clustering of GLUT4, TUG, and RUVBL2 protein levels correlate with myosin heavy chain isoform pattern in skeletal muscles, but AS160 and TBC1D1 levels do not. J. Appl. Physiol. 2011, 111, 1106–1117. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.; Mueller, B.; Ploegh, H.L.; Schlieker, C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell 2009, 36, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Ishibashi, T.; Ogawa, S.; Hashiguchi, Y.; Inoue, Y.; Udo, H.; Ohzono, H.; Kato, A.; Minakami, R.; Sugiyama, H. A novel protein specifically interacting with Homer2 regulates ubiquitin-proteasome systems. J. Biochem. 2005, 137, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Park, J.K.; Jeong, J.; Jeon, H.; Yoon, J.B.; Kim, E.E.; Lee, K.J. Complex of Fas-associated factor 1 (FAF1) with valosin-containing protein (VCP)-Npl4-Ufd1 and polyubiquitinated proteins promotes endoplasmic reticulum-associated degradation (ERAD). J. Biol. Chem. 2013, 288, 6998–7011. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Yin, C.; Doong, H.; Fang, S.; Peterhoff, C.; Nixon, R.A.; Monteiro, M.J. Characterization of erasin (UBXD2): A new ER protein that promotes ER-associated protein degradation. J. Cell Sci. 2006, 119 Pt 19, 4011–4024. [Google Scholar] [CrossRef] [PubMed]
- Madsen, L.; Kriegenburg, F.; Vala, A.; Best, D.; Prag, S.; Hofmann, K.; Seeger, M.; Adams, I.R.; Hartmann-Petersen, R. The tissue-specific Rep8/UBXD6 tethers p97 to the endoplasmic reticulum membrane for degradation of misfolded proteins. PLoS ONE 2011, 6, e25061. [Google Scholar] [CrossRef] [PubMed]
- Schuberth, C.; Buchberger, A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 2005, 7, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.; Sane, S.; Freeling, J.L.; Wang, H.; Zhang, D.; Rezvani, K. Nucleocytoplasmic Translocation of UBXN2A Is Required for Apoptosis during DNA Damage Stresses in Colon Cancer Cells. J. Cancer 2015, 6, 1066–1078. [Google Scholar] [CrossRef] [PubMed]
- LaLonde, D.P.; Bretscher, A. The UBX protein SAKS1 negatively regulates endoplasmic reticulum-associated degradation and p97-dependent degradation. J. Biol. Chem. 2011, 286, 4892–4901. [Google Scholar] [CrossRef] [PubMed]
- Araki, K.; Nagata, K. Protein folding and quality control in the ER. Cold Spring Harb. Perspect. Biol. 2012, 4, a015438. [Google Scholar] [CrossRef] [PubMed]
- Buchberger, A.; Howard, M.J.; Proctor, M.; Bycroft, M. The UBX domain: A widespread ubiquitin-like module. J. Mol. Biol. 2001, 307, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Meyer, H.H.; Rapoport, T.A. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 2001, 414, 652–656. [Google Scholar] [CrossRef] [PubMed]
- Schuberth, C.; Buchberger, A. UBX domain proteins: Major regulators of the AAA ATPase CDC48/p97. Cell. Mol. Life Sci. 2008, 65, 2360–2371. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.Y.; Kang, W.; Lee, S.Y.; Yang, J.K. Crystallization and preliminary X-ray crystallographic analysis of the N domain of p97/VCP in complex with the UBX domain of FAF1. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66 Pt 1, 41–43. [Google Scholar] [CrossRef] [PubMed]
- Yeung, H.O.; Kloppsteck, P.; Niwa, H.; Isaacson, R.L.; Matthews, S.; Zhang, X.; Freemont, P.S. Insights into adaptor binding to the AAA protein p97. Biochem. Soc. Trans. 2008, 36, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Alberts, S.M.; Sonntag, C.; Schafer, A.; Wolf, D.H. UBX4 modulates cdc48 activity and influences degradation of misfolded proteins of the endoplasmic reticulum. J. Biol. Chem. 2009, 284, 16082–16089. [Google Scholar] [CrossRef] [PubMed]
- Nagahama, M.; Ohnishi, M.; Kawate, Y.; Matsui, T.; Miyake, H.; Yuasa, K.; Tani, K.; Tagaya, M.; Tsuji, A. UBXD1 is a VCP-interacting protein that is involved in ER-associated degradation. Biochem. Biophys. Res. Commun. 2009, 382, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.D.; Liu, Y.; Bentivoglio, C.M.; Barlowe, C. SEL1P/UBX2P participates in a distinct CDC48P-dependent endoplasmic reticulum-associated degradation pathway. Traffic 2006, 7, 1213–1223. [Google Scholar] [CrossRef] [PubMed]
- Rezvani, K.; Teng, Y.; Pan, Y.; Dani, J.A.; Lindstrom, J.; Garcia Gras, E.A.; McIntosh, J.M.; de Biasi, M. UBXD4, a UBX-containing protein, regulates the cell surface number and stability of α3-containing nicotinic acetylcholine receptors. J. Neurosci. 2009, 29, 6883–6896. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Rezvani, K.; de Biasi, M. UBXN2A regulates nicotinic receptor degradation by modulating the E3 ligase activity of CHIP. Biochem. Pharmacol. 2015, 97, 518–530. [Google Scholar] [CrossRef] [PubMed]
- Meyer, H.; Bug, M.; Bremer, S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 2012, 14, 117–123. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Zhu, Q.; Wani, G.; Sharma, N.; Wani, A.A. Valosin-containing Protein (VCP)/p97 Segregase Mediates Proteolytic Processing of Cockayne Syndrome Group B (CSB) in Damaged Chromatin. J. Biol. Chem. 2016, 291, 7396–7408. [Google Scholar] [CrossRef] [PubMed]
- Kloppsteck, P.; Ewens, C.A.; Forster, A.; Zhang, X.; Freemont, P.S. Regulation of p97 in the ubiquitin-proteasome system by the UBX protein-family. Biochim. Biophys. Acta 2012, 1823, 125–129. [Google Scholar] [CrossRef] [PubMed]
- Haines, D.S. p97-containing complexes in proliferation control and cancer: Emerging culprits or guilt by association? Genes Cancer 2010, 1, 753–763. [Google Scholar] [CrossRef] [PubMed]
- Moriguchi, S.; Maehara, Y.; Korenaga, D.; Sugimachi, K.; Nose, Y. Risk factors which predict pattern of recurrence after curative surgery for patients with advanced gastric cancer. Surg. Oncol. 1992, 1, 341–346. [Google Scholar] [CrossRef]
- Motoori, M.; Takemasa, I.; Yano, M.; Saito, S.; Miyata, H.; Takiguchi, S.; Fujiwara, Y.; Yasuda, T.; Doki, Y.; Kurokawa, Y.; et al. Prediction of recurrence in advanced gastric cancer patients after curative resection by gene expression profiling. Int. J. Cancer 2005, 114, 963–968. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.G.; Kim, H.J.; Dlugosz, A.A.; Ellison, D.W.; Gilbertson, R.J.; Alvarez-Buylla, A. Dual and opposing roles of primary cilia in medulloblastoma development. Nat. Med. 2009, 15, 1062–1065. [Google Scholar] [CrossRef] [PubMed]
- Menzl, I.; Lebeau, L.; Pandey, R.; Hassounah, N.B.; Li, F.W.; Nagle, R.; Weihs, K.; McDermott, K.M. Loss of primary cilia occurs early in breast cancer development. Cilia 2014, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Schraml, P.; Frew, I.J.; Thoma, C.R.; Boysen, G.; Struckmann, K.; Krek, W.; Moch, H. Sporadic clear cell renal cell carcinoma but not the papillary type is characterized by severely reduced frequency of primary cilia. Mod. Pathol. 2009, 22, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Seeley, E.S.; Carriere, C.; Goetze, T.; Longnecker, D.S.; Korc, M. Pancreatic cancer and precursor pancreatic intraepithelial neoplasia lesions are devoid of primary cilia. Cancer Res. 2009, 69, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Bhogaraju, S.; Engel, B.D.; Lorentzen, E. Intraflagellar transport complex structure and cargo interactions. Cilia 2013, 2, 10. [Google Scholar] [CrossRef] [PubMed]
- Raman, M.; Sergeev, M.; Garnaas, M.; Lydeard, J.R.; Huttlin, E.L.; Goessling, W.; Shah, J.V.; Harper, J.W. Systematic proteomics of the VCP-UBXD adaptor network identifies a role for UBXN10 in regulating ciliogenesis. Nat. Cell Biol. 2015, 17, 1356–1369. [Google Scholar] [CrossRef] [PubMed]
- Sane, S.; Abdullah, A.; Nelson, M.E.; Wang, H.; Chauhan, S.C.; Newton, S.S.; Rezvani, K. Structural studies of UBXN2A and mortalin interaction and the putative role of silenced UBXN2A in preventing response to chemotherapy. Cell Stress Chaperones 2016, 21, 313–326. [Google Scholar] [CrossRef] [PubMed]
- Dores-Silva, P.R.; Barbosa, L.R.; Ramos, C.H.; Borges, J.C. Human mitochondrial Hsp70 (mortalin): Shedding light on ATPase activity, interaction with adenosine nucleotides, solution structure and domain organization. PLoS ONE 2015, 10, e0117170. [Google Scholar] [CrossRef] [PubMed]
- Black, J.D.; Rezvani, K. Heat Shock Protein 70s as Potential Molecular Targets for Colon Cancer Therapeutics. Curr. Med. Chem. 2016. [Google Scholar] [CrossRef]
- Gestl, E.E.; Anne Bottger, S. Cytoplasmic sequestration of the tumor suppressor p53 by a heat shock protein 70 family member, mortalin, in human colorectal adenocarcinoma cell lines. Biochem. Biophys. Res. Commun. 2012, 423, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Kaul, S.C.; Duncan, E.L.; Englezou, A.; Takano, S.; Reddel, R.R.; Mitsui, Y.; Wadhwa, R. Malignant transformation of NIH3T3 cells by overexpression of mot-2 protein. Oncogene 1998, 17, 907–911. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.; Kaul, Z.; Yoon, A.R.; Liu, Y.; Yaguchi, T.; Na, Y.; Ahn, H.M.; Gao, R.; Choi, I.K.; Yun, C.O.; et al. Identification and functional characterization of nuclear mortalin in human carcinogenesis. J. Biol. Chem. 2014, 289, 24832–24844. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.J.; Lee, N.P.; Kaul, S.C.; Lan, F.; Poon, R.T.; Wadhwa, R.; Luk, J.M. Mortalin-p53 interaction in cancer cells is stress dependent and constitutes a selective target for cancer therapy. Cell Death Differ. 2011, 6, 1046–1056. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.J.; Lee, N.P.; Kaul, S.C.; Lan, F.; Poon, R.T.; Wadhwa, R.; Luk, J.M. Induction of mutant p53-dependent apoptosis in human hepatocellular carcinoma by targeting stress protein mortalin. Int. J. Cancer 2011, 129, 1806–1814. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.K.; Hong, S.K.; Veeranki, S.; Karkhanis, M.; Starenki, D.; Plaza, J.A.; Park, J.I. A mortalin/HSPA9-mediated switch in tumor-suppressive signaling of Raf/MEK/extracellular signal-regulated kinase. Mol. Cell. Biol. 2013, 33, 4051–4067. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Takano, S.; Robert, M.; Yoshida, A.; Nomura, H.; Reddel, R.R.; Mitsui, Y.; Kaul, S.C. Inactivation of tumor suppressor p53 by mot-2, a HSP70 family member. J. Biol. Chem. 1998, 273, 29586–29591. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Takano, S.; Kaur, K.; Deocaris, C.C.; Pereira-Smith, O.M.; Reddel, R.R.; Kaul, S.C. Upregulation of mortalin/mthsp70/Grp75 contributes to human carcinogenesis. Int. J. Cancer 2006, 118, 2973–2980. [Google Scholar] [CrossRef] [PubMed]
- Deocaris, C.C.; Lu, W.J.; Kaul, S.C.; Wadhwa, R. Druggability of mortalin for cancer and neuro-degenerative disorders. Curr. Pharm. Des. 2013, 19, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Pilzer, D.; Saar, M.; Koya, K.; Fishelson, Z. Mortalin inhibitors sensitize K562 leukemia cells to complement-dependent cytotoxicity. Int. J. Cancer 2010, 126, 1428–1435. [Google Scholar] [CrossRef] [PubMed]
- Wadhwa, R.; Taira, K.; Kaul, S. Can mortalin be a candidate target for cancertherapy? Cancer Ther. 2003, 1, 173–178. [Google Scholar]
- Sane, S.; Abdullah, A.; Boudreau, D.A.; Autenried, R.K.; Gupta, B.K.; Wang, X.; Wang, H.; Schlenker, E.H.; Zhang, D.; Telleria, C.; et al. Ubiquitin-like (UBX)-domain-containing protein, UBXN2A, promotes cell death by interfering with the p53-Mortalin interactions in colon cancer cells. Cell Death Dis. 2014, 5, e1118. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, A.; Sane, S.; Branick, K.A.; Freeling, J.L.; Wang, H.; Zhang, D.; Rezvani, K. A plant alkaloid, veratridine, potentiates cancer chemosensitivity by UBXN2A-dependent inhibition of an oncoprotein, mortalin-2. Oncotarget 2015, 6, 23561–23581. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Li, Y.F.; El-Gamil, M.; Rosenberg, S.A.; Robbins, P.F. Identification of a colorectal tumor-associated antigen (COA-1) recognized by CD4(+) T lymphocytes. Cancer Res. 2003, 63, 6735–6743. [Google Scholar] [PubMed]
- Quintero, M.; Mackenzie, N.; Brennan, P.A. Hypoxia-inducible factor 1 (HIF-1) in cancer. Eur. J. Surg. Oncol. 2004, 30, 465–468. [Google Scholar] [CrossRef] [PubMed]
- Giaccia, A.; Siim, B.G.; Johnson, R.S. HIF-1 as a target for drug development. Nat. Rev. Drug Discov. 2003, 2, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Parada, L.F. Neurofibromin, a tumor suppressor in the nervous system. Exp. Cell Res. 2001, 264, 19–28. [Google Scholar] [CrossRef] [PubMed]
- Phan, V.T.; Ding, V.W.; Li, F.; Chalkley, R.J.; Burlingame, A.; McCormick, F. The RasGAP proteins Ira2 and neurofibromin are negatively regulated by Gpb1 in yeast and ETEA in humans. Mol. Cell. Biol. 2010, 30, 2264–2279. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, K.; Bucher, P. The UBA domain: A sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem. Sci. 1996, 21, 172–173. [Google Scholar] [CrossRef]
- Yuan, X.; Simpson, P.; McKeown, C.; Kondo, H.; Uchiyama, K.; Wallis, R.; Dreveny, I.; Keetch, C.; Zhang, X.; Robinson, C.; et al. Structure, dynamics and interactions of p47, a major adaptor of the AAA ATPase, p97. EMBO J. 2004, 23, 1463–1473. [Google Scholar] [CrossRef] [PubMed]
- Soukenik, M.; Diehl, A.; Leidert, M.; Sievert, V.; Bussow, K.; Leitner, D.; Labudde, D.; Ball, L.J.; Lechner, A.; Nagler, D.K.; et al. The SEP domain of p47 acts as a reversible competitive inhibitor of cathepsin L. FEBS Lett. 2004, 576, 358–362. [Google Scholar] [CrossRef] [PubMed]
- Besche, H.C.; Haas, W.; Gygi, S.P.; Goldberg, A.L. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 2009, 48, 2538–2549. [Google Scholar] [CrossRef] [PubMed]
- Kondo, H.; Rabouille, C.; Newman, R.; Levine, T.P.; Pappin, D.; Freemont, P.; Warren, G. p47 is a cofactor for p97-mediated membrane fusion. Nature 1997, 388, 75–78. [Google Scholar] [PubMed]
- Meyer, H.H.; Wang, Y.; Warren, G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and UFD1-NPL4. EMBO J. 2002, 21, 5645–5652. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Jokitalo, E.; Lindman, M.; Jackman, M.; Kano, F.; Murata, M.; Zhang, X.; Kondo, H. The localization and phosphorylation of p47 are important for Golgi disassembly-assembly during the cell cycle. J. Cell. Biol. 2003, 161, 1067–1079. [Google Scholar] [CrossRef] [PubMed]
- Uchiyama, K.; Kondo, H. p97/p47-Mediated biogenesis of Golgi and ER. J. Biochem. 2005, 137, 115–119. [Google Scholar] [CrossRef] [PubMed]
- Roy, L.; Bergeron, J.J.; Lavoie, C.; Hendriks, R.; Gushue, J.; Fazel, A.; Pelletier, A.; Morre, D.J.; Subramaniam, V.N.; Hong, W.; et al. Role of p97 and syntaxin 5 in the assembly of transitional endoplasmic reticulum. Mol. Biol. Cell 2000, 11, 2529–2542. [Google Scholar] [CrossRef] [PubMed]
- Hetzer, M.; Meyer, H.H.; Walther, T.C.; Bilbao-Cortes, D.; Warren, G.; Mattaj, I.W. Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat. Cell Biol. 2001, 3, 1086–1091. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Copley, R.R.; Doerks, T.; Ponting, C.P.; Bork, P. SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000, 28, 231–234. [Google Scholar] [CrossRef] [PubMed]
- Herszenyi, L.; Plebani, M.; Carraro, P.; de Paoli, M.; Roveroni, G.; Rugge, M.; Cardin, R.; Naccarato, R.; Farinati, F. Role and behavior of cathepsin B and cathepsin L in gastric cancer. Orv. Hetil. 1995, 136, 1315–1318. [Google Scholar] [PubMed]
- Mohamed, M.M.; Sloane, B.F. Cysteine cathepsins: Multifunctional enzymes in cancer. Nat. Rev. Cancer 2006, 6, 764–775. [Google Scholar] [CrossRef] [PubMed]
- Thomssen, C.; Schmitt, M.; Goretzki, L.; Oppelt, P.; Pache, L.; Dettmar, P.; Janicke, F.; Graeff, H. Prognostic value of the cysteine proteases cathepsins B and cathepsin L in human breast cancer. Clin. Cancer Res. 1995, 1, 741–746. [Google Scholar] [PubMed]
- Foekens, J.A.; Kos, J.; Peters, H.A.; Krasovec, M.; Look, M.P.; Cimerman, N.; Meijer-van Gelder, M.E.; Henzen-Logmans, S.C.; van Putten, W.L.; Klijn, J.G. Prognostic significance of cathepsins B and L in primary human breast cancer. J. Clin. Oncol. 1998, 16, 1013–1021. [Google Scholar] [PubMed]
- Levicar, N.; Dewey, R.A.; Daley, E.; Bates, T.E.; Davies, D.; Kos, J.; Pilkington, G.J.; Lah, T.T. Selective suppression of cathepsin L by antisense cDNA impairs human brain tumor cell invasion in vitro and promotes apoptosis. Cancer Gene Ther. 2003, 10, 141–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krepela, E. Cysteine proteinases in tumor cell growth and apoptosis. Neoplasma 2001, 48, 332–349. [Google Scholar] [PubMed]
- Karin, M. Nuclear factor-κB in cancer development and progression. Nature 2006, 441, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Chaturvedi, M.M.; Sung, B.; Yadav, V.R.; Kannappan, R.; Aggarwal, B.B. NF-κB addiction and its role in cancer: ‘One size does not fit all’. Oncogene 2011, 30, 1615–1630. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Lee, S. NF-κB in cellular senescence and cancer treatment. Mol. Cells 2014, 37, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Wu, P.; Zhao, L.; Huang, L.; Zhang, Z.; Zhao, S.; Huang, J. NF-κB Expression and Outcomes in Solid Tumors: A Systematic Review and Meta-Analysis. Medicine 2015, 94, e1687. [Google Scholar] [CrossRef] [PubMed]
- Le Page, C.; Koumakpayi, I.H.; Lessard, L.; Mes-Masson, A.M.; Saad, F. EGFR and Her-2 regulate the constitutive activation of NF-κB in PC-3 prostate cancer cells. Prostate 2005, 65, 130–140. [Google Scholar] [CrossRef] [PubMed]
- Birkenkamp, K.U.; Geugien, M.; Schepers, H.; Westra, J.; Lemmink, H.H.; Vellenga, E. Constitutive NF-κB DNA-binding activity in AML is frequently mediated by a Ras/PI3-K/PKB-dependent pathway. Leukemia 2004, 18, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Jeon, Y.T.; Kim, S.H.; Song, Y.S. NF-κB as a potential molecular target for cancer therapy. BioFactors 2007, 29, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Hawke, N.; Baldwin, A.S. NF-κB and IKK as therapeutic targets in cancer. Cell Death Differ. 2006, 13, 738–747. [Google Scholar] [CrossRef] [PubMed]
- Hayden, M.S.; Ghosh, S. Shared principles in NF-κB signaling. Cell 2008, 132, 344–362. [Google Scholar] [CrossRef] [PubMed]
- Hadian, K.; Griesbach, R.A.; Dornauer, S.; Wanger, T.M.; Nagel, D.; Metlitzky, M.; Beisker, W.; Schmidt-Supprian, M.; Krappmann, D. NF-κB essential modulator (NEMO) interaction with linear and lys-63 ubiquitin chains contributes to NF-κB activation. J. Biol. Chem. 2011, 286, 26107–26117. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.J.; Conze, D.B.; Li, T.; Srinivasula, S.M.; Ashwell, J.D. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation [corrected]. Nat. Cell Biol. 2006, 8, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Chiaravalli, J.; Fontan, E.; Fsihi, H.; Coic, Y.M.; Baleux, F.; Veron, M.; Agou, F. Direct inhibition of NF-κB activation by peptide targeting the NOA ubiquitin binding domain of NEMO. Biochem. Pharmacol. 2011, 82, 1163–1174. [Google Scholar] [CrossRef] [PubMed]
- Safa, A.R. c-FLIP, a master anti-apoptotic regulator. Exp. Oncol. 2012, 34, 176–184. [Google Scholar] [PubMed]
- Fulda, S.; Vucic, D. Targeting IAP proteins for therapeutic intervention in cancer. Nat. Rev. Drug Discov. 2012, 11, 109–124. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Oyama, M.; Kozuka-Hata, H.; Han, X.; Tanaka, Y.; Gohda, J.; Inoue, J. p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO. Nat. Commun. 2012, 3, 1061. [Google Scholar] [CrossRef] [PubMed]
- Adham, I.M.; Khulan, J.; Held, T.; Schmidt, B.; Meyer, B.I.; Meinhardt, A.; Engel, W. Fas-associated factor (FAF1) is required for the early cleavage-stages of mouse embryo. Mol. Hum. Reprod. 2008, 14, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Menges, C.W.; Altomare, D.A.; Testa, J.R. FAS-associated factor 1 (FAF1): Diverse functions and implications for oncogenesis. Cell Cycle 2009, 8, 2528–2534. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.H.; Kang, W.; Suh, S.W.; Yang, J.K. Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain. Proteins 2011, 79, 2583–2587. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Park, J.K.; Lee, J.J.; Choi, Y.S.; Ryu, K.S.; Kim, J.H.; Kim, E.; Lee, K.J.; Jeon, Y.H.; Kim, E.E. Structure and interaction of ubiquitin-associated domain of human Fas-associated factor 1. Protein Sci. 2009, 18, 2265–2276. [Google Scholar] [CrossRef] [PubMed]
- Song, E.J.; Yim, S.H.; Kim, E.; Kim, N.S.; Lee, K.J. Human Fas-associated factor 1, interacting with ubiquitinated proteins and valosin-containing protein, is involved in the ubiquitin-proteasome pathway. Mol. Cell. Biol. 2005, 25, 2511–2524. [Google Scholar] [CrossRef] [PubMed]
- Beauparlant, P.; Kwan, I.; Bitar, R.; Chou, P.; Koromilas, A.E.; Sonenberg, N.; Hiscott, J. Disruption of IκBα regulation by antisense RNA expression leads to malignant transformation. Oncogene 1994, 9, 3189–3197. [Google Scholar] [PubMed]
- Carrasco, D.; Perez, P.; Lewin, A.; Bravo, R. IκBα overexpression delays tumor formation in v-rel transgenic mice. J. Exp. Med. 1997, 186, 279–288. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.M.; Nozaki, S.; Shortle, N.H.; Bhat-Nakshatri, P.; Newton, T.R.; Rice, S.; Gelfanov, V.; Boswell, S.H.; Goulet, R.J., Jr.; Sledge, G.W., Jr.; et al. Paclitaxel sensitivity of breast cancer cells with constitutively active NF-κB is enhanced by IkappaBalpha super-repressor and parthenolide. Oncogene 2000, 19, 4159–4169. [Google Scholar] [CrossRef] [PubMed]
- Milacic, V.; Banerjee, S.; Landis-Piwowar, K.R.; Sarkar, F.H.; Majumdar, A.P.; Dou, Q.P. Curcumin inhibits the proteasome activity in human colon cancer cells in vitro and in vivo. Cancer Res. 2008, 68, 7283–7292. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Moon, H.S.; Chung, H.W. The expression of FAS-associated factor 1 and heat shock protein 70 in ovarian cancer. Obstet. Gynecol. Sci. 2014, 57, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Chu, K.; Niu, X.; Williams, L.T. A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc. Natl. Acad. Sci. USA 1995, 92, 11894–11898. [Google Scholar] [CrossRef] [PubMed]
- Ryu, S.W.; Lee, S.J.; Park, M.Y.; Jun, J.I.; Jung, Y.K.; Kim, E. Fas-associated factor 1, FAF1, is a member of Fas death-inducing signaling complex. J. Biol. Chem. 2003, 278, 24003–24010. [Google Scholar] [CrossRef] [PubMed]
- Jang, M.S.; Sul, J.W.; Choi, B.J.; Lee, S.J.; Suh, J.H.; Kim, N.S.; Kim, W.H.; Lim, D.S.; Lee, C.W.; Kim, E. Negative feedback regulation of Aurora-A via phosphorylation of Fas-associated factor-1. J. Biol. Chem. 2008, 283, 32344–32351. [Google Scholar] [CrossRef] [PubMed]
- Kinoshita, T.; Kondoh, C.; Hasegawa, M.; Imamura, R.; Suda, T. Fas-associated factor 1 is a negative regulator of PYRIN-containing Apaf-1-like protein 1. Int. Immunol. 2006, 18, 1701–1706. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Jang, H.D.; Lee, S.Y.; Lee, K.J.; Kim, E. Fas-associated factor-1 inhibits nuclear factor-κB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-κB. J. Biol. Chem. 2004, 279, 2544–2549. [Google Scholar] [CrossRef] [PubMed]
- Park, M.Y.; Moon, J.H.; Lee, K.S.; Choi, H.I.; Chung, J.; Hong, H.J.; Kim, E. FAF1 suppresses IκB kinase (IKK) activation by disrupting the IKK complex assembly. J. Biol. Chem. 2007, 282, 27572–27577. [Google Scholar] [CrossRef] [PubMed]
- Altomare, D.A.; Menges, C.W.; Pei, J.; Zhang, L.; Skele-Stump, K.L.; Carbone, M.; Kane, A.B.; Testa, J.R. Activated TNF-α/NF-κB signaling via down-regulation of Fas-associated factor 1 in asbestos-induced mesotheliomas from ARF knockout mice. Proc. Natl. Acad. Sci. USA 2009, 106, 3420–3425. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Gasparri, F.; Galvani, A.; Nici, L.; Darnowski, J.W.; Barbone, D.; Fennell, D.A.; Gaudino, G.; Porta, C.; Mutti, L. Bortezomib inhibits nuclear factor-κB dependent survival and has potent in vivo activity in mesothelioma. Clin. Cancer Res. 2007, 13, 5942–5951. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, N.; Morisaki, T.; Hashizume, K.; Yao, T.; Tsuneyoshi, M.; Noshiro, H.; Nakamura, K.; Yamanaka, T.; Uchiyama, A.; Tanaka, M.; et al. Nuclear factor-κB p65 (RelA) transcription factor is constitutively activated in human gastric carcinoma tissue. Clin. Cancer Res. 2001, 7, 4136–4142. [Google Scholar] [PubMed]
- Kim, H.J.; Song, E.J.; Lee, Y.S.; Kim, E.; Lee, K.J. Human Fas-associated factor 1 interacts with heat shock protein 70 and negatively regulates chaperone activity. J. Biol. Chem. 2005, 280, 8125–8133. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.J.; Kim, Y.M.; Jeong, J.; Bae, D.S.; Lee, K.J. Ubiquitin-Associated (UBA) Domain in Human Fas Associated Factor 1 Inhibits Tumor Formation by Promoting Hsp70 Degradation. PLoS ONE 2012, 7, e40361. [Google Scholar] [CrossRef] [PubMed]
- Amantini, C.; Mosca, M.; Nabissi, M.; Lucciarini, R.; Caprodossi, S.; Arcella, A.; Giangaspero, F.; Santoni, G. Capsaicin-induced apoptosis of glioma cells is mediated by TRPV1 vanilloid receptor and requires p38 MAPK activation. J. Neurochem. 2007, 102, 977–990. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, C.; Shin, C.Y.; Hwang, S.W.; Yang, Y.D.; Shim, W.S.; Park, M.Y.; Kim, E.; Kim, M.; Kim, B.M.; et al. TRPV1 recapitulates native capsaicin receptor in sensory neurons in association with Fas-associated factor 1. J. Neurosci. 2006, 26, 2403–2412. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.K.; Basu, S. Fas-associated factor 1 is a negative regulator in capsaicin induced cancer cell apoptosis. Cancer Lett. 2010, 287, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Deveraux, Q.L.; Roy, N.; Stennicke, H.R.; van Arsdale, T.; Zhou, Q.; Srinivasula, S.M.; Alnemri, E.S.; Salvesen, G.S.; Reed, J.C. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998, 17, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Mahoney, D.J.; Cheung, H.H.; Mrad, R.L.; Plenchette, S.; Simard, C.; Enwere, E.; Arora, V.; Mak, T.W.; Lacasse, E.C.; Waring, J.; et al. Both cIAP1 and cIAP2 regulate TNFα-mediated NF-κB activation. Proc. Natl. Acad. Sci. USA 2008, 105, 11778–11783. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, M.J.; Milutinovic, S.; Dickson, K.M.; Ho, W.C.; Boudreault, A.; Durkin, J.; Gillard, J.W.; Jaquith, J.B.; Morris, S.J.; Barker, P.A. cIAP1 and cIAP2 facilitate cancer cell survival by functioning as E3 ligases that promote RIP1 ubiquitination. Mol. Cell 2008, 30, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Gyrd-Hansen, M.; Meier, P. IAPs: From caspase inhibitors to modulators of NF-κB, inflammation and cancer. Nat. Rev. Cancer 2010, 10, 561–574. [Google Scholar] [CrossRef] [PubMed]
- Fulda, S. Targeting inhibitor of apoptosis proteins for cancer therapy: A double-edge sword? J. Clin. Oncol. 2014, 32, 3190–3191. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.B.; Tan, B.; Mu, R.; Chang, Y.; Wu, M.; Tu, H.Q.; Zhang, Y.C.; Guo, S.S.; Qin, X.H.; Li, T.; et al. Ubiquitin-associated domain-containing ubiquitin regulatory X (UBX) protein UBXN1 is a negative regulator of nuclear factor κB (NF-κB) signaling. J. Biol. Chem. 2015, 290, 10395–10405. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.J. Ubiquitination in signaling to and activation of IKK. Immunol. Rev. 2012, 246, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Wajant, H.; Scheurich, P. TNFR1-induced activation of the classical NF-κB pathway. FEBS J. 2011, 278, 862–876. [Google Scholar] [CrossRef] [PubMed]
- De Almagro, M.C.; Vucic, D. The inhibitor of apoptosis (IAP) proteins are critical regulators of signaling pathways and targets for anti-cancer therapy. Exp. Oncol. 2012, 34, 200–211. [Google Scholar] [PubMed]
- Morris, J.R.; Pangon, L.; Boutell, C.; Katagiri, T.; Keep, N.H.; Solomon, E. Genetic analysis of BRCA1 ubiquitin ligase activity and its relationship to breast cancer susceptibility. Hum. Mol. Genet. 2006, 15, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Wu-Baer, F.; Ludwig, T.; Baer, R. The UBXN1 protein associates with autoubiquitinated forms of the BRCA1 tumor suppressor and inhibits its enzymatic function. Mol. Cell. Biol. 2010, 30, 2787–2798. [Google Scholar] [CrossRef] [PubMed]
- Denicourt, C.; Saenz, C.C.; Datnow, B.; Cui, X.S.; Dowdy, S.F. Relocalized p27Kip1 tumor suppressor functions as a cytoplasmic metastatic oncogene in melanoma. Cancer Res. 2007, 67, 9238–9243. [Google Scholar] [CrossRef] [PubMed]
- Kossatz, U.; Malek, N.P. p27: Tumor suppressor and oncogene ...? Cell Res. 2007, 17, 832–833. [Google Scholar] [CrossRef] [PubMed]
- Devoy, A.; Soane, T.; Welchman, R.; Mayer, R.J. The ubiquitin-proteasome system and cancer. Essays Biochem. 2005, 41, 187–203. [Google Scholar] [CrossRef] [PubMed]
- Dou, Q.P.; Zonder, J.A. Overview of proteasome inhibitor-based anti-cancer therapies: Perspective on bortezomib and second generation proteasome inhibitors versus future generation inhibitors of ubiquitin-proteasome system. Curr. Cancer Drug Targets 2014, 14, 517–536. [Google Scholar] [CrossRef] [PubMed]
- Frezza, M.; Schmitt, S.; Dou, Q.P. Targeting the ubiquitin-proteasome pathway: An emerging concept in cancer therapy. Curr. Top. Med. Chem. 2011, 11, 2888–2905. [Google Scholar] [CrossRef] [PubMed]
- Mackay, H.; Hedley, D.; Major, P.; Townsley, C.; Mackenzie, M.; Vincent, M.; Degendorfer, P.; Tsao, M.S.; Nicklee, T.; Birle, D.; et al. A phase II trial with pharmacodynamic endpoints of the proteasome inhibitor bortezomib in patients with metastatic colorectal cancer. Clin. Cancer Res. 2005, 11, 5526–5533. [Google Scholar] [CrossRef] [PubMed]
- Avet-Loiseau, H.; Fonseca, R.; Siegel, D.; Dimopoulos, M.A.; Spicka, I.; Masszi, T.; Hajek, R.; Rosinol, L.; Goranova-Marinova, V.; Mihaylov, G.; et al. Carfilzomib significantly improves the progression free survival of high-risk patients in multiple myeloma. Blood 2016. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. Next-generation proteasome blockers promise safer cancer therapy. Nat. Med. 2012, 18, 7. [Google Scholar] [CrossRef] [PubMed]
- Ruschak, A.M.; Slassi, M.; Kay, L.E.; Schimmer, A.D. Novel proteasome inhibitors to overcome bortezomib resistance. J. Natl. Cancer Inst. 2011, 103, 1007–1017. [Google Scholar] [CrossRef] [PubMed]
- Alfaraj, W.A.; Cachia, D.; Tummala, S.; Thomas, S.K.; Manasanch, E.E. Severe peripheral neuropathy following carfilzomib, rituximab, and dexamethasone for initial treatment of Waldenstrom’s macroglobulinemia. Ann. Hematol. 2016, 95, 347–348. [Google Scholar] [CrossRef] [PubMed]
- Jhaveri, K.D.; Wanchoo, R. Carfilzomib-induced nephrotoxcity. Kidney Int. 2015, 88, 199–200. [Google Scholar] [CrossRef] [PubMed]
- Tjionas, H.; Gupta, A.K. Heart failure secondary to carfilzomib-induced heart block in multiple myeloma patients. J. Oncol. Pharm. Pract. 2016. [Google Scholar] [CrossRef] [PubMed]
- Pal, A.; Young, M.A.; Donato, N.J. Emerging potential of therapeutic targeting of ubiquitin-specific proteases in the treatment of cancer. Cancer Res. 2014, 74, 4955–4966. [Google Scholar] [CrossRef] [PubMed]
- Anderson, D.J.; le Moigne, R.; Djakovic, S.; Kumar, B.; Rice, J.; Wong, S.; Wang, J.; Yao, B.; Valle, E.; Kiss von Soly, S.; et al. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer Cell 2015, 28, 653–665. [Google Scholar] [CrossRef] [PubMed]




© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rezvani, K. UBXD Proteins: A Family of Proteins with Diverse Functions in Cancer. Int. J. Mol. Sci. 2016, 17, 1724. https://doi.org/10.3390/ijms17101724
Rezvani K. UBXD Proteins: A Family of Proteins with Diverse Functions in Cancer. International Journal of Molecular Sciences. 2016; 17(10):1724. https://doi.org/10.3390/ijms17101724
Chicago/Turabian StyleRezvani, Khosrow. 2016. "UBXD Proteins: A Family of Proteins with Diverse Functions in Cancer" International Journal of Molecular Sciences 17, no. 10: 1724. https://doi.org/10.3390/ijms17101724
APA StyleRezvani, K. (2016). UBXD Proteins: A Family of Proteins with Diverse Functions in Cancer. International Journal of Molecular Sciences, 17(10), 1724. https://doi.org/10.3390/ijms17101724