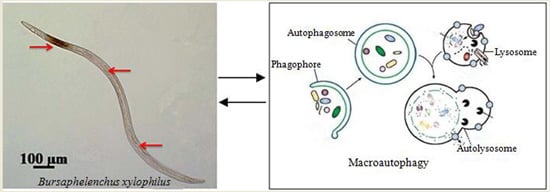
Identification of Autophagy in the Pine Wood Nematode Bursaphelenchus xylophilus and the Molecular Characterization and Functional Analysis of Two Novel Autophagy-Related Genes, BxATG1 and BxATG8
Abstract
:1. Introduction
2. Results
2.1. Qualitative Identification of Autophagy in B. xylophilus by Transmission Electron Microscopy (TEM)
2.2. Autophagy-Related Gene Homologues in B. xylophilus
2.3. In Situ Hybridization (ISH) for the Localization of BxATG8 in B. xylophilus
2.4. Detection of RNAi Efficiency
2.5. Effect of RNAi on B. xylophilus Reproduction on Fungal Mats
3. Discussion
4. Materials and Methods
4.1. B. xylophilus Growth Conditions and Experimental Organisms
4.2. TEM as Tool to Study Autophagy in B. xylophilus
4.3. RNA Isolation and cDNA Synthesis of B. xylophilus
4.4. Homology-Based Cloning of Partial BxATG1 and BxATG8 Sequences from B. xylophilus
4.5. Full-Length cDNA Cloning of BxATG1 and BxATG8 from B. xylophilus
4.6. Cloning and Sequencing of BxAtg1 and BxAtg8
4.7. ISH
4.8. BxATG1 and BxATG8 Interference Using Double-Stranded RNA
4.9. Analysis of Reproduction of B. xylophilus after RNAi
4.10. Quantitative Reverse Transcription PCR (qRT-PCR)
4.11. Statistical Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mamiya, Y. Pathology of the pine wilt disease caused by Bursaphelenchus xylophilus. Annu. Rev. Phytopathol. 1983, 21, 201–220. [Google Scholar] [CrossRef] [PubMed]
- Kanzaki, N.; Futai, K. A PCR primer set for determination of phylogenetic relationships of Bursaphelenchus species within the xylophilus group. Nematology 2002, 4, 35–41. [Google Scholar] [CrossRef]
- Ceng, H.R.; Lin, M.S.; Ni, W.Q.; Fang, Z.D. First report of pine wilt disease from Pinus thunbergii Parl in Nanjing. Pak. J. Nematol. 1983, 4, 1–5. [Google Scholar]
- Mota, M.M.; Braasch, H.; Bravo, M.A.; Burgermeister, W.; Metge, K.; Sousa, E. First report of Bursaphelenchus xylophilus in Portugal and in Europe. Nematology 1999, 1, 727–734. [Google Scholar] [CrossRef]
- Yang, B.J.; Wang, Q.L. Distribution of the pinewood nematode in China and susceptibility of some Chinese and exotic pines to the nematode. Can. J. For. Res. 1989, 19, 1527–1530. [Google Scholar]
- Braasch, H.; Tomiczek, C.; Metge, K.; Hoyer, U.; Burgermeister, W.; Wulfert, I.; Schonfeld, U. Records of Bursaphelenchus spp. (Nematoda, Parasitaphelenchidae) in coniferous timber imported from the Asian part of Russia. For. Pathol. 2001, 31, 129–140. [Google Scholar] [CrossRef]
- Son, J.A.; Hogetsu, T.; Moon, Y.S. The process of epithelial cell death in Pinus thunbergii caused by the pine wood nematode, Bursaphelenchus xylophilus. Nematology 2014, 16, 663–668. [Google Scholar] [CrossRef]
- Zhao, B.G.; Wang, H.L.; Han, S.F.; Zheng, M.H. Distribution and pathogenicity of bacteria species carried by Bursaphelenchus xylophilus in China. Nematology 2003, 5, 899–906. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, C.Y.; Fang, Z.M.; Zhang, D.L.; Liu, L.; Lee, M.R.; Li, Z.; Li, J.J.; Sung, C.K. Advances in research of pathogenic mechanism of pine wilt disease. Afr. J. Microbiol. Res. 2010, 4, 437–442. [Google Scholar]
- Kikuchi, T.; Cotton, J.A.; Dalzell, J.J.; Hasegawa, K.; Kanzaki, N.; McVeigh, P.; Takanashi, T.; Tsai, I.J.; Assefa, S.A.; Cock, P.J.A.; et al. Genomic insights into the origin of parasitism in the emerging plant pathogen Bursaphelenchus xylophilus. PLoS Pathog. 2011, 7, 1059–1074. [Google Scholar] [CrossRef] [PubMed]
- Ogura, N.; Nakashima, T. In vitro occurrence of dispersal fourth stage juveniles in Bursaphelenchus xylophilus co-incubated with Monochamus alternatus. Jpn. J. Nematol. 2002, 32, 53–59. [Google Scholar]
- Meléndez, A.; Talloczy, Z.; Seaman, M.; Eskelinen, E.L.; Hall, D.H.; Levine, B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003, 301, 1387–1391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chang, J.T.; Guo, B.; Hansen, M.; Jia, K.; Kovács, A.L.; Kumsta, C.; Lapierre, L.R.; Legouis, R.; Lin, L.; et al. Guidelines for monitoring autophagy in Caenorhabditis elegans. Autophagy 2015, 11, 9–27. [Google Scholar] [PubMed]
- Levine, B.; Klionsky, D.J. Development by self-digestion: Molecular mechanisms and biological functions of autophagy. Dev. Cell 2004, 6, 463–477. [Google Scholar] [CrossRef]
- Meléndez, A.; Levine, B. Autophagy in C. elegans. NCBI [Internet]. Available online: http://www.ncbi.nlm.nih.gov/books/NBK116074/ (accessed on 21 August 2015).
- Klionsky, D.J. The molecular machinery of autophagy: Unanswered questions. J. Cell Sci. 2005, 118, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005, 12, 1535–1541. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.N.; Jeong, T.J. Overview of autophagy in plant cells. J. Life Sci. 2014, 24, 209–217. [Google Scholar] [CrossRef]
- Liu, X.H.; Lu, J.P.; Zhang, L.; Dong, B.; Min, H.; Lin, F.C. Involvement of a Magnaporthe grisea serine/threonine kinase gene, MgATG1, in appressorium turgor and pathogenesis. Eukaryot. Cell 2007, 6, 997–1005. [Google Scholar] [CrossRef] [PubMed]
- Tindwa, H.; Jo, Y.H.; Patnaik, B.B.; Lee, Y.S.; Kang, S.S.; Han, Y.S. Molecular cloning and characterization of autophagy-related gene TmATG8 in Listeria-invaded hemocytes of Tenebrio molitor. Dev. Comp. Immunol. 2015, 51, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Fernández, J.M.F.; Ortega, A.G.; Cruz, R.R.; Camberos, E.P.; Álvarez, Á.H.; Velázquez, M.M. Molecular cloning and characterization of two novel autophagy-related genes belonging to the ATG8 family from the cattle tick Rhipicephalus (Boophilus) microplus (Acari: Ixodidae). Exp. Appl. Acarol. 2014, 64, 533–542. [Google Scholar] [CrossRef] [PubMed]
- Valent, B.; Farrall, L.; Chumley, F.G. Magnaporthe grisea genes for pathogenicity and virulence identified through a series of backcrosses. Genetics 1991, 127, 87–101. [Google Scholar] [PubMed]
- Ylä-Anttila, P.; Vihinen, H.; Jokitalo, E.; Eskelinen, E.L. Monitoring autophagy by electron microscopy in Mammalian cells. Methods Enzymol. 2009, 452, 143–164. [Google Scholar] [PubMed]
- Spellman, P.T.; Sherlock, G.; Zhang, M.Q.; Iyer, V.R.; Anders, K.; Eisen, M.B.; Brown, P.O.; Botstein, D.; Futcher, B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 1998, 9, 3273–3297. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.Y.; Tooze, S.A. Evolution of Atg1 function and regulation. Autophagy 2009, 5, 758–765. [Google Scholar] [CrossRef] [PubMed]
- Shpilka, T.; Weidberg, H.; Pietrokovski, S.; Elazar, Z. Atg8: An autophagy-related ubiquitin-like protein family. Genome Biol. 2011, 12. [Google Scholar] [CrossRef] [PubMed]
- Ryter, S.W.; Cloonan, S.M.; Choi, A.M.K. Autophagy: A critical regulator of cellular metabolism and homeostasis. Mol. Cells 2013, 36, 7–16. [Google Scholar] [CrossRef] [PubMed]
- Vandekerckhove, T.T.M.; Coomans, A.; Cornelis, K.; Cornelis, K.; Baert, P.; Gillis, M. Use of the Verrucomicrobia-specific probe EUB338-III and fluorescent in situ hybridization for detection of “Candidatus Xiphinematobacter” cells in nematode hosts. Appl. Environ. Microbiol. 2002, 68, 3121–3125. [Google Scholar] [CrossRef] [PubMed]
- De Boer, J.M.; Yan, Y.; Smant, G.; Davis, E.L.; Baum, T.J. In-situ hybridization to messenger RNA in Heterodera glycines. J. Nematol. 1998, 30, 309–312. [Google Scholar] [PubMed]
- Wang, F.; Wang, Z.Y.; Li, D.L.; Chen, Q.L. Identification and characterization of a Bursaphelenchus xylophilus (Aphelenchida: Aphelenchoididae) thermotolerance-related gene: Bx-hsp90. Int. J. Mol. Sci. 2012, 13, 8819–8833. [Google Scholar] [CrossRef] [PubMed]
- Cheong, H.; Klionsky, D.J. Biochemical methods to monitor autophagy-related processes in yeast. Methods Enzymol. 2008, 451, 1–26. [Google Scholar] [PubMed]
- Navale, R.; Allanki, A.D.; Sijwali, P.S. Characterization of the autophagy marker protein Atg8 reveals atypical features of autophagy in Plasmodium falciparum. PLoS ONE 2014, 9, e113220. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Yoshimori, T.; Levine, B. Methods in mammalian autophagy research. Cell 2010, 140, 313–326. [Google Scholar] [CrossRef] [PubMed]
- De Boer, J.M.; Smant, G.; Goverse, A.; Davis, E.L.; Ovremars, H.A.; Pomp, H.; Gent-Pelzer, M.V.; Zilverentant, J.F.; Stokkermans, J.P.W.G.; Hussey, R.S.; et al. Secretory granule proteins from the subventral esophageal glands of the potato cyst nematode identified by monoclonal antibodies to a protein fraction from second-stage juvenile. Mol. Plant Microbe Interact. 1996, 9, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, T.; Shibuya, H.; Aikawa, T.; Jones, J.T. Cloning and characterization of pectate lyases expressed in the esophageal gland of the pine wood nematode Bursaphelenchus xylophilus. Mol. Plant Microbe Interact. 2006, 19, 280–287. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.X. Plant Nematology; Chinese Agricultural Publishing House: Beijing, China, 2001; p. 11. [Google Scholar]
- Asakura, M.; Ninomiya, S.; Sugimoto, M.; Oku, M.; Yamashita, S.I.; Okuno, T.; Sakai, Y.; Takano, Y. Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbicular. Plant Cell 2009, 21, 1291–1304. [Google Scholar] [CrossRef] [PubMed]
- Bryant, B.; Raikhel, A.S. Programmed autophagy in the fat body of Aedes aegypti is required to maintain egg maturation cycles. PLoS ONE 2011, 6, e25502. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.Q.; Montgomery, M.K.; Kostas1, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Wesley, S.V.; Helliwell, C.A.; Smith, N.A.; Wang, M.B.; Rouse, D.T.; Liu, Q.; Gooding, P.S.; Singh, S.P.; Abbott, D.; Stoutjesdijk, P.A.; et al. Construct design for efficient. effective and high-throughput gene silencing in plants. Plant J. 2001, 27, 581–590. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.H.; Wang, H.I.; Lu, H.C.; Chen, C.E.; Chen, H.H.; Yeh, H.H.; Tang, C.Y. An efficient RNA interference screening strategy for gene functional analysis. BMC Genom. 2012, 13. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.B.; Lu, Q.; Liang, J.; Zhang, X.Y. Functional analysis of the cellulose gene of the pine wood nematode, Bursaphelenchus xylophilus, using RNA interference. Genet. Mol. Res. 2011, 10, 1931–1941. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Koh, Y.H.; Moon, Y.S.; Lee, S.H. Molecular properties of a venom allergen-like protein suggest a parasitic function in the pinewood nematode Bursaphelenchus xylophilus. Int. J. Parasitol. 2012, 42, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.Y.; Dai, S.M.; Xiao, L.; Xie, B.Y. Influence of cellulase gene knockdown by dsRNA interference on the development and reproduction of the pine wood nematode, Bursaphelenchus xylophilus. Nematology 2010, 12, 225–233. [Google Scholar] [CrossRef]
- Xu, X.L.; Wu, X.Q.; Ye, J.R.; Huang, L. Molecular characterization and functional analysis of three pathogenesis-related Cytochrome P450 genes from Bursaphelenchus xylophilus (Tylenchida: Aphelenchoidoidea). Int. J. Mol. Sci. 2015, 16, 5216–5234. [Google Scholar] [CrossRef] [PubMed]
- Aladzsity, I.; Tóth, M.L.; Sigmond, T.; Szabó, E.; Bicsák, B.; Barna, J.; Regős, Á.; Orosz, L.; Kovács, A.L.; Vellai, T. Autophagy genes unc-51 and bec-1 are required for normal cell size in Caenorhabditis elegan. Genetics 2007, 177, 655–660. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Wu, X.Q.; Zhou, A.D. Bacterial diversity and community structure in the pine wood nematode Bursaphelenchus xylophilus and B. mucronatus with different virulence by high-throughput sequencing of the 16S rDNA. PLoS ONE 2015, 10, e0137386. [Google Scholar] [CrossRef] [PubMed]
- Viglierchio, D.R.; Schmitt, R.V. On the methodology of nematode extraction from field samples: Baermann funnel modifications. J. Nematol. 1983, 15, 438–444. [Google Scholar] [PubMed]
- Martinet, W.; Timmermans, J.P.; de Meyer, G.R.Y. Methods to assess autophagy in situ—Transmission electron microscopy versus immunohistochemistry. Methods Enzymol. 2014, 543, 89–114. [Google Scholar] [PubMed]
- Consensus-degenerate hybrid oligonucleotide primers. Available online: http://blocks.fhcrc.org/codehop.html (accessed on 25 August 2015).
- Urwin, P.E.; Lilley, C.J.; Atkinson, H.J. Ingestion of double-stranded RNA by preparasitic juvenile cyst nematodes leads to RNA interference. Mol. Plant Microbe Interact. 2002, 15, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yamada, T.; Sakaue, D.; Suzuki, K. Variations in life history parameters and their influence on rate of population increase of different pathogenic isolates of the pine wood nematode, Bursaphelenchus xylophilus. Nematology 2005, 7, 459–467. [Google Scholar] [CrossRef]







| Name of Primers | Sequence (5′–3′) |
|---|---|
| F-BxATG1 | GGGCCGAGCAGCTGGTNGTNTAYGT |
| R-BxATG1 | CACAGCTCGATGGCGTGNYKRTACAT |
| F-BxATG8 | ACTTTGAGAAGCGTCGTG |
| R-BxATG8 | TGTGGAATGACATTGTTGAC |
| GSP1-1 | TCTTGTGATGGCTCAACGAC |
| GSP1-2 | CTCGTTCTCAAGAGCTGGCT |
| GSP1-3 | TGGAGATGGCTGAAGAGTCG |
| GSP1-4 | CGTTGAGCCATCACAAGACT |
| GSP8-1 | GTCGTGCTGAAGGTGAGAAGAT |
| GSP8-2 | TGAAGGTGAGAAGATCCGTCGCAAGT |
| GSP8-3 | ATAAAGCTGTCCCATCGTGGTCGT |
| GSP8-4 | TCAGAGGGAACCAGATACTT |
| BxATG1-T7I-F | TAATACGACTCACTATAGGGAAGGCAGAAATCGGACA |
| BxATG1-I-R | AATCGGCTCATGGAAAA |
| BxATG1-I-F | AAGGCAGAAATCGGACA |
| BxATG1-T7I-R | TAATACGACTCACTATAGGGAATCGGCTCATGGAAAA |
| BxATG8-T7I-F | TAATACGACTCACTATAGGGAACCCAAGTTTGAGACCT |
| BxATG8-I-R | CGAAAACACTACAATAAGA |
| BxATG8-I-F | AACCCAAGTTTGAGACCT |
| BxATG8-T7I-R | TAATACGACTCACTATAGGGCGAAAACACTACAATAAGA |
| Actin F | GCAACACGGAGTTCGTTGTAGA |
| Actin R | GTATCGTCACCAACTGGGATGA |
| qBxATG1-F | AGAGTGTTGGGTGAGGGA |
| qBxATG1-R | CTCGGCATTGGTACATTATA |
| qBxATG8-F | GTCAACGATGTCATTCCCCA |
| qBxATG8-R | AACTGATCACTCTTCGGCGG |
| M13F(-47) | CGCCAGGGTTTTCCCAGTCACGAC |
| M13R(-48) | AGCGGATAACAATTTCACACAGGA |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, L.-N.; Wu, X.-Q.; Ye, J.-R.; Xue, Q. Identification of Autophagy in the Pine Wood Nematode Bursaphelenchus xylophilus and the Molecular Characterization and Functional Analysis of Two Novel Autophagy-Related Genes, BxATG1 and BxATG8. Int. J. Mol. Sci. 2016, 17, 279. https://doi.org/10.3390/ijms17030279
Deng L-N, Wu X-Q, Ye J-R, Xue Q. Identification of Autophagy in the Pine Wood Nematode Bursaphelenchus xylophilus and the Molecular Characterization and Functional Analysis of Two Novel Autophagy-Related Genes, BxATG1 and BxATG8. International Journal of Molecular Sciences. 2016; 17(3):279. https://doi.org/10.3390/ijms17030279
Chicago/Turabian StyleDeng, Li-Na, Xiao-Qin Wu, Jian-Ren Ye, and Qi Xue. 2016. "Identification of Autophagy in the Pine Wood Nematode Bursaphelenchus xylophilus and the Molecular Characterization and Functional Analysis of Two Novel Autophagy-Related Genes, BxATG1 and BxATG8" International Journal of Molecular Sciences 17, no. 3: 279. https://doi.org/10.3390/ijms17030279
APA StyleDeng, L.-N., Wu, X.-Q., Ye, J.-R., & Xue, Q. (2016). Identification of Autophagy in the Pine Wood Nematode Bursaphelenchus xylophilus and the Molecular Characterization and Functional Analysis of Two Novel Autophagy-Related Genes, BxATG1 and BxATG8. International Journal of Molecular Sciences, 17(3), 279. https://doi.org/10.3390/ijms17030279