New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies
Abstract
:1. Introduction
1.1. Origin of Extracellular miRNAs
1.2. Cell–Cell Communication (Hormone-Like Molecules)
2. miRNAs as Cancer Biomarkers
2.1. let-7 Family (let-7a, -7b, -7c, -7e, -7f, -7i)
2.2. miR-10b
2.3. miR-16
2.4. The miR-17~92 Cluster
2.5. miR-21
2.6. The miR-29 Family (miR-29a, -29b, and -29c)
2.7. The miR-30 Family (miR-30a, -30b, -30c, -30d, and -30e)
2.8. The miR-34 Family (miR-34a, -34b, and -34c)
2.9. The miR-125 Family (miR-125a and -125b)
2.10. miR-155
2.11. The miR-200 Family (miR-200a, -200b, -200c, -141, and -429)
2.12. miR-210
2.13. miR-221/-222
2.14. miR-375
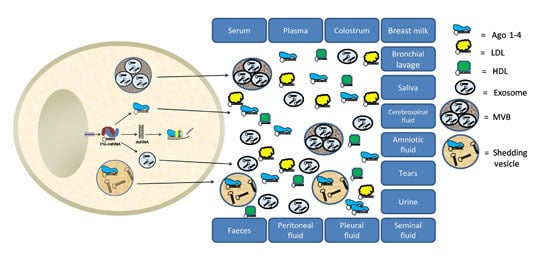
3. Extracellular miRNAs in Other Biological Fluids
4. Discussion
Challenges in Studying cfmiRNA
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization. Available online: http://www.who.int/mediacentre/factsheets/fs297/en/ (accessed on 10 January 2016).
- Pathak, A.K.; Bhutani, M.; Kumar, S.; Mohan, A.; Guleria, R. Circulating cell-free DNA in plasma/serum of lung cancer patients as a potential screening and prognostic tool. Clin. Chem. 2006, 52, 1833–1842. [Google Scholar] [PubMed]
- Mandel, P.; Metais, P. Les acides nucléiques du plasma sanguin chez l’homme. C. R. Acad. Sci. Paris 1948, 142, 241–243. [Google Scholar]
- Tan, E.M.; Schur, P.H.; Carr, R.I.; Kunkel, H.G. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Investig. 1966, 45, 1732–1740. [Google Scholar] [CrossRef] [PubMed]
- Ayala, W.; Moore, L.V.; Hess, E.L. The purple color reaction given by diphenylamine reagent. I. With normal and rheumatic fever sera. J. Clin. Investig. 1951, 30, 781–785. [Google Scholar] [CrossRef] [PubMed]
- Leon, S.A.; Shapiro, B.; Sklaroff, D.M.; Yaros, M.J. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977, 37, 646–650. [Google Scholar] [PubMed]
- Vasioukhin, V.; Anker, P.; Maurice, P.; Lyautey, J.; Lederrey, C.; Stroun, M. Point mutations of the N-ras gene in the blood plasma DNA of patients with myelodysplastic syndrome or acute myelogenous leukaemia. Br. J. Haematol. 1994, 86, 774–779. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble normal and mutated DNA sequences from single-copy genes in human blood. Cancer Epidemiol. Biomark. Prev. 1994, 3, 67–71. [Google Scholar]
- Lo, K.W.; Lo, Y.M.; Leung, S.F.; Tsang, Y.S.; Chan, L.Y.; Johnson, P.J.; Hjelm, N.M.; Lee, J.C.; Huang, D.P. Analysis of cell-free Epstein-Barr virus associated RNA in the plasma of patients with nasopharyngeal carcinoma. Clin. Chem. 1999, 45, 1292–1294. [Google Scholar] [PubMed]
- Lawrie, C.H. MicroRNA expression in lymphoma. Expert Opin. Biol. Ther. 2007, 7, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.N. MicroRNA biogenesis: Coordinated cropping and dicing. Nat. Rev. Mol. Cell Biol. 2005, 6, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Burwinkel, B. Extracellular miRNAs: The mystery of their origin and function. Trends Biochem. Sci. 2012, 37, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Bueso-Ramos, C.; Ferdin, J.; Lopez-Berestein, G.; Sood, A.K.; Calin, G.A. MicroRNAs in body fluids—The mix of hormones and biomarkers. Nat. Rev. Clin. Oncol. 2011, 8, 467–477. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W. Circulating microRNA biomarker studies: Pitfalls and potential solutions. Clin. Chem. 2015, 61, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Weiz, L.; Langheinz, A.; Burwinkel, B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011, 39, 7223–7233. [Google Scholar] [CrossRef] [PubMed]
- Laterza, O.F.; Lim, L.; Garrett-Engele, P.W.; Vlasakova, K.; Muniappa, N.; Tanaka, W.K.; Johnson, J.M.; Sina, J.F.; Fare, T.L.; Sistare, F.D.; et al. Plasma microRNAs as sensitive and specific biomarkers of tissue injury. Clin. Chem. 2009, 55, 1977–1983. [Google Scholar] [CrossRef] [PubMed]
- Corsten, M.F.; Dennert, R.; Jochems, S.; Kuznetsova, T.; Devaux, Y.; Hofstra, L.; Wagner, D.R.; Staessen, J.A.; Heymans, S.; Schroen, B. Circulating microRNA-208b and microRNA-499 reflect myocardial damage in cardiovascular disease. Circ. Cardiovasc. Genet. 2010, 3, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Gholamin, S.; Pasdar, A.; Khorrami, M.S.; Mirzaei, H.; Mirzaei, H.R.; Salehi, R.; Ferns, G.A.; Ghayour-Mobarhan, M.; Avan, A. The potential for circulating microRNAs in the diagnosis of myocardial infarction: A novel approach to disease diagnosis and treatment. Curr. Pharm. Des. 2016, 22, 397–403. [Google Scholar] [CrossRef] [PubMed]
- Sayed, A.S.; Xia, K.; Yang, T.L.; Peng, J. Circulating microRNAs: A potential role in diagnosis and prognosis of acute myocardial infarction. Dis. Markers 2013, 35, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Anker, P.; Mulcahy, H.; Stroun, M. Circulating nucleic acids in plasma and serum as a noninvasive investigation for cancer: Time for large-scale clinical studies? Int. J. Cancer 2003, 103, 149–152. [Google Scholar] [CrossRef] [PubMed]
- Hunter, M.P.; Ismail, N.; Zhang, X.; Aguda, B.D.; Lee, E.J.; Yu, L.; Xiao, T.; Schafer, J.; Lee, M.L.; Schmittgen, T.D.; et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS ONE 2008, 3, e3694. [Google Scholar] [CrossRef] [PubMed]
- Ismail, N.; Wang, Y.; Dakhlallah, D.; Moldovan, L.; Agarwal, K.; Batte, K.; Shah, P.; Wisler, J.; Eubank, T.D.; Tridandapani, S.; et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 2013, 121, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Zhang, S.; Weber, J.; Baxter, D.; Galas, D.J. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 2010, 38, 7248–7259. [Google Scholar] [CrossRef] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, J.D.; Chevillet, J.R.; Kroh, E.M.; Ruf, I.K.; Pritchard, C.C.; Gibson, D.F.; Mitchell, P.S.; Bennett, C.F.; Pogosova-Agadjanyan, E.L.; Stirewalt, D.L.; et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. USA 2011, 108, 5003–5008. [Google Scholar] [CrossRef] [PubMed]
- Gallo, A.; Tandon, M.; Alevizos, I.; Illei, G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS ONE 2012, 7, e30679. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Gonzalez, S.; Sanchez-Cabo, F.; Gonzalez, M.A.; Bernad, A.; Sanchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef] [PubMed]
- Baulcombe, D.C. RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol. 1996, 32, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Turchinovich, A.; Samatov, T.R.; Tonevitsky, A.G.; Burwinkel, B. Circulating miRNAs: Cell–cell communication function? Front. Genet. 2013, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Thery, C. Exosomes: Secreted vesicles and intercellular communications. F1000 Biol. Rep. 2011, 3, 15. [Google Scholar] [CrossRef] [PubMed]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Wurdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef] [PubMed]
- Yuan, A.; Farber, E.L.; Rapoport, A.L.; Tejada, D.; Deniskin, R.; Akhmedov, N.B.; Farber, D.B. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS ONE 2009, 4, e4722. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Li, J.; Chen, W.X.; Cai, Y.Q.; Yu, D.D.; Zhong, S.L.; Zhao, J.H.; Zhou, J.W.; Tang, J.H. Exosomes decrease sensitivity of breast cancer cells to adriamycin by delivering microRNAs. Tumour Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- De Souza, P.S.; Faccion, R.S.; Bernardo, P.S.; Maia, R.C. Membrane microparticles: Shedding new light into cancer cell communication. Cancer Res. Clin. Oncol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Maushagen, R.; Pries, R.; Wollenberg, B. Chemotherapy with paclitaxel leads to microRNA release. HNO 2015, 63, 792–796. [Google Scholar] [CrossRef] [PubMed]
- Hannafon, B.N.; Carpenter, K.J.; Berry, W.L.; Janknecht, R.; Dooley, W.C.; Ding, W.Q. Exosome-mediated microRNA signaling from breast cancer cells is altered by the anti-angiogenesis agent docosahexaenoic acid (DHA). Mol. Cancer 2015, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Gai, C.; Carpanetto, A.; Deregibus, M.C.; Camussi, G. Extracellular vesicle-mediated modulation of angiogenesis. Histol. Histopathol. 2015, 34, 11708. [Google Scholar]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Soneji, S.; Marafioti, T.; Cooper, C.D.; Palazzo, S.; Paterson, J.C.; Cattan, H.; Enver, T.; Mager, R.; Boultwood, J.; et al. MicroRNA expression distinguishes between germinal center B cell–like and activated B cell–like subtypes of diffuse large B cell lymphoma. Int. J. Cancer 2007, 121, 1156–1161. [Google Scholar] [CrossRef] [PubMed]
- Mo, M.H.; Chen, L.; Fu, Y.; Wang, W.; Fu, S.W. Cell-free Circulating miRNA Biomarkers in Cancer. J. Cancer 2012, 3, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef] [PubMed]
- Kelly, B.D.; Miller, N.; Sweeney, K.J.; Durkan, G.C.; Rogers, E.; Walsh, K.; Kerin, M.J. A Circulating MicroRNA Signature as a Biomarker for Prostate Cancer in a High Risk Group. J. Clin. Med. 2015, 4, 1369–1379. [Google Scholar] [CrossRef] [PubMed]
- Ogata-Kawata, H.; Izumiya, M.; Kurioka, D.; Honma, Y.; Yamada, Y.; Furuta, K.; Gunji, T.; Ohta, H.; Okamoto, H.; Sonoda, H.; et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS ONE 2014, 9, e92921. [Google Scholar]
- Ghanbari, R.; Mosakhani, N.; Sarhadi, V.K.; Armengol, G.; Nouraee, N.; Mohammadkhani, A.; Khorrami, S.; Arefian, E.; Paryan, M.; Malekzadeh, R.; et al. Simultaneous Underexpression of let-7a-5p and let-7f-5p microRNAs in Plasma and Stool Samples from Early Stage Colorectal Carcinoma. Biomark. Cancer 2015, 7, 39–48. [Google Scholar] [PubMed]
- Zuo, Z.; Calin, G.A.; de Paula, H.M.; Medeiros, L.J.; Fernandez, M.H.; Shimizu, M.; Garcia-Manero, G.; Bueso-Ramos, C.E. Circulating microRNAs let-7a and miR-16 predict progression-free survival and overall survival in patients with myelodysplastic syndrome. Blood 2011, 118, 413–415. [Google Scholar] [CrossRef] [PubMed]
- Tsujiura, M.; Ichikawa, D.; Komatsu, S.; Shiozaki, A.; Takeshita, H.; Kosuga, T.; Konishi, H.; Morimura, R.; Deguchi, K.; Fujiwara, H.; et al. Circulating microRNAs in plasma of patients with gastric cancers. Br. J. Cancer 2010, 102, 1174–1179. [Google Scholar] [CrossRef] [PubMed]
- Heegaard, N.H.; Schetter, A.J.; Welsh, J.A.; Yoneda, M.; Bowman, E.D.; Harris, C.C. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int. J. Cancer 2012, 130, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Hung, C.H.; Hu, T.H.; Lu, S.N.; Kuo, F.Y.; Chen, C.H.; Wang, J.H.; Huang, C.M.; Lee, C.M.; Lin, C.Y.; Yen, Y.H.; et al. Circulating microRNAs as biomarkers for diagnosis of early hepatocellular carcinoma associated with hepatitis B virus. Int. J. Cancer 2016, 138, 714–720. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.W.; Bae, H.S.; Song, J.Y.; Lee, J.K.; Lee, N.W.; Kim, T.; Lee, K.W. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patients. Int. J. Gynecol. Cancer 2013, 23, 673–679. [Google Scholar] [CrossRef] [PubMed]
- Dou, H.; Wang, Y.; Su, G.; Zhao, S. Decreased plasma let-7c and miR-152 as noninvasive biomarker for non-small-cell lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 9291–9298. [Google Scholar] [PubMed]
- Li, X.X.; Gao, S.Y.; Wang, P.Y.; Zhou, X.; Li, Y.J.; Yu, Y.; Yan, Y.F.; Zhang, H.H.; Lv, C.J.; Zhou, H.H.; et al. Reduced expression levels of let-7c in human breast cancer patients. Oncol. Lett. 2015, 9, 1207–1212. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Xu, Q.; Sun, L.P.; Dong, Q.G.; He, C.Y.; Yuan, Y. Expression of serum let-7c, let-7i, and let-7f microRNA with its target gene, pepsinogen C, in gastric cancer and precancerous disease. Tumour Biol. 2015, 36, 3337–3343. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Almhanna, K.; Chen, W.; Philip, P.A.; Sarkar, F.H. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am. J. Transl. Res. 2010, 3, 28–47. [Google Scholar] [PubMed]
- Kubiczkova, L.; Kryukov, F.; Slaby, O.; Dementyeva, E.; Jarkovsky, J.; Nekvindova, J.; Radova, L.; Greslikova, H.; Kuglik, P.; Vetesnikova, E.; et al. Circulating serum microRNAs as novel diagnostic and prognostic biomarkers for multiple myeloma and monoclonal gammopathy of undetermined significance. Haematologica 2014, 99, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Liu, Y.; Wang, J.; Guo, Z.; Zhang, Q.; Yu, F.; Zhang, Y.; Huang, K.; Li, Y.; Song, E.; et al. Circulating microRNA profiles as potential biomarkers for diagnosis of papillary thyroid carcinoma. J. Clin. Endocrinol. Metab. 2012, 97, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Zhang, L.; Zhao, Y.; Yang, D.; Song, F.; Wen, Y.; Hao, Q.; Hu, Z.; Zhang, W.; Chen, K. Plasma miRNAs as diagnostic and prognostic biomarkers for ovarian cancer. PLoS ONE 2013, 8, e77853. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Garcia, V.; Zaballos, A.; Provencio, M.; Lombardia, L.; Almonacid, L.; Garcia, J.M.; Dominguez, G.; Pena, C.; Diaz, R.; et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Ge, W.; Yu, D.C.; Li, Q.G.; Chen, X.; Zhang, C.Y.; Ding, Y.T. Expression of serum miR-16, let-7f, and miR-21 in patients with hepatocellular carcinoma and their clinical significances. Clin. Lab. 2014, 60, 427–434. [Google Scholar] [PubMed]
- Huang, J.; Wu, J.; Li, Y.; Li, X.; Yang, T.; Yang, Q.; Jiang, Y. Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. BioMed Res. Int. 2014, 2014, 364316. [Google Scholar] [CrossRef] [PubMed]
- Langhe, R.; Norris, L.; Saadeh, F.A.; Blackshields, G.; Varley, R.; Harrison, A.; Gleeson, N.; Spillane, C.; Martin, C.; O'Donnell, D.M.; et al. A novel serum microRNA panel to discriminate benign from malignant ovarian disease. Cancer Lett. 2015, 356, 628–636. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, S.; Giordano, A.; Gao, H.; Cohen, E.N.; Tin, S.; Wu, Q.; Garza, R.J.; Debeb, B.G.; Alvarez, R.H.; Valero, V.; et al. High serum miR-19a levels are associated with inflammatory breast cancer and are predictive of favorable clinical outcome in patients with metastatic HER2+ inflammatory breast cancer. PLoS ONE 2014, 9, e83113. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Rack, B.; Muller, V.; Janni, W.; Pantel, K.; Schwarzenbach, H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010, 12, R90. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012, 14, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Roth, C.; Kasimir-Bauer, S.; Pantel, K.; Schwarzenbach, H. Screening for circulating nucleic acids and caspase activity in the peripheral blood as potential diagnostic tools in lung cancer. Mol. Oncol. 2011, 5, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Yao, Y.; Meng, F.; Qian, X.; Jiang, X.; Li, X.; Gao, Z.; Gao, L. Predictive Value of Serum miR-10b, miR-29c, and miR-205 as Promising Biomarkers in Esophageal Squamous Cell Carcinoma Screening. Medicine (Baltimore) 2015, 94, e1558. [Google Scholar] [CrossRef] [PubMed]
- Maclellan, S.A.; Lawson, J.; Baik, J.; Guillaud, M.; Poh, C.F.; Garnis, C. Differential expression of miRNAs in the serum of patients with high-risk oral lesions. Cancer Med. 2012, 1, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Lodes, M.J.; Caraballo, M.; Suciu, D.; Munro, S.; Kumar, A.; Anderson, B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS ONE 2009, 4, e6229. [Google Scholar] [CrossRef] [PubMed]
- Mahn, R.; Heukamp, L.C.; Rogenhofer, S.; von Ruecker, A.; Muller, S.C.; Ellinger, J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology 2011, 77, 1265.e9–1265.e16. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Dong, J.; Wang, L.E.; Ma, H.; Liu, J.; Zhao, Y.; Tang, J.; Chen, X.; Dai, J.; Wei, Q.; et al. Serum microRNA profiling and breast cancer risk: The use of miR-484/191 as endogenous controls. Carcinogenesis 2012, 33, 828–834. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhang, K.; Liu, L.H.; Ouyang, Y.; Guo, H.B.; Zhang, H.; Bu, J.; Xiao, T. MicroRNA screening identifies circulating microRNAs as potential biomarkers for osteosarcoma. Oncol. Lett. 2015, 10, 1662–1668. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Song, Y.; Zhang, C.; Zhi, X.; Fu, H.; Ma, Y.; Chen, Y.; Pan, F.; Wang, K.; Ni, J.; et al. Circulating MiR-16–5p and MiR-19b-3p as Two Novel Potential Biomarkers to Indicate Progression of Gastric Cancer. Theranostics 2015, 5, 733–745. [Google Scholar] [CrossRef]
- Wang, H.; Wang, L.; Wu, Z.; Sun, R.; Jin, H.; Ma, J.; Liu, L.; Ling, R.; Yi, J.; Wang, L.; et al. Three dysregulated microRNAs in serum as novel biomarkers for gastric cancer screening. Med. Oncol. 2014, 31, 298. [Google Scholar] [CrossRef] [PubMed]
- Li, B.X.; Yu, Q.; Shi, Z.L.; Li, P.; Fu, S. Circulating microRNAs in esophageal squamous cell carcinoma: Association with locoregional staging and survival. Int. J. Clin. Exp. Med. 2015, 8, 7241–7250. [Google Scholar]
- Hirajima, S.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Konishi, H.; Shiozaki, A.; Morimura, R.; Tsujiura, M.; Nagata, H.; Kawaguchi, T.; et al. Clinical impact of circulating miR-18a in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 2013, 108, 1822–1829. [Google Scholar] [CrossRef]
- Tsujiura, M.; Komatsu, S.; Ichikawa, D.; Shiozaki, A.; Konishi, H.; Takeshita, H.; Moriumura, R.; Nagata, H.; Kawaguchi, T.; Hirajima, S.; et al. Circulating miR-18a in plasma contributes to cancer detection and monitoring in patients with gastric cancer. Gastric Cancer 2015, 18, 271–279. [Google Scholar] [CrossRef]
- Kodahl, A.R.; Lyng, M.B.; Binder, H.; Cold, S.; Gravgaard, K.; Knoop, A.S.; Ditzel, H.J. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: A case control study. Mol. Oncol. 2014, 8, 874–883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yau, T.O.; Wu, C.W.; Dong, Y.; Tang, C.M.; Ng, S.S.; Chan, F.K.; Sung, J.J.; Yu, J. microRNA-221 and microRNA-18a identification in stool as potential biomarkers for the non-invasive diagnosis of colorectal carcinoma. Br. J. Cancer 2014, 111, 1765–1771. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Grossmann, K.F.; Cassidy, P.B.; Yang, C.H.; Fan, M.; Kopelovich, L.; Leachman, S.A.; Pfeffer, L.M. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J. Clin. Med. 2015, 4, 2012–2027. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, M.; Ma, F.; Luo, Y.; Cai, R.; Wang, L.; Xu, N.; Xu, B. Circulating miR-19a and miR-205 in serum may predict the sensitivity of luminal A subtype of breast cancer patients to neoadjuvant chemotherapy with epirubicin plus paclitaxel. PLoS ONE 2014, 9, e104870. [Google Scholar] [CrossRef] [PubMed]
- Sochor, M.; Basova, P.; Pesta, M.; Dusilkova, N.; Bartos, J.; Burda, P.; Pospisil, V.; Stopka, T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014, 14, 448. [Google Scholar] [CrossRef]
- Feng, Y.; Liu, J.; Kang, Y.; He, Y.; Liang, B.; Yang, P.; Yu, Z. miR-19a acts as an oncogenic microRNA and is up-regulated in bladder cancer. J. Exp. Clin. Cancer Res. 2014, 33, 67. [Google Scholar] [CrossRef] [PubMed]
- Matsumura, T.; Sugimachi, K.; Iinuma, H.; Takahashi, Y.; Kurashige, J.; Sawada, G.; Ueda, M.; Uchi, R.; Ueo, H.; Takano, Y.; et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br. J. Cancer 2015, 113, 275–281. [Google Scholar] [CrossRef] [PubMed]
- Hao, M.; Zang, M.; Wendlandt, E.; Xu, Y.; An, G.; Gong, D.; Li, F.; Qi, F.; Zhang, Y.; Yang, Y.; et al. Low serum miR-19a expression as a novel poor prognostic indicator in multiple myeloma. Int. J. Cancer 2015, 136, 1835–1844. [Google Scholar] [CrossRef]
- Lin, Q.; Chen, T.; Lin, Q.; Lin, G.; Lin, J.; Chen, G.; Guo, L. Serum miR-19a expression correlates with worse prognosis of patients with non-small cell lung cancer. J. Surg. Oncol. 2013, 107, 767–771. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Cao, Y.; He, Z.; He, J.; Hu, C.; Duan, H.; Jiang, J. Serum levels of miR-19b and miR-146a as prognostic biomarkers for non-small cell lung cancer. Tohoku J. Exp. Med. 2014, 232, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Hruby, G.W.; McKiernan, J.M.; Gurvich, I.; Lipsky, M.J.; Benson, M.C.; Santella, R.M. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate 2012, 72, 1469–1477. [Google Scholar] [CrossRef] [PubMed]
- Geng, Q.; Fan, T.; Zhang, B.; Wang, W.; Xu, Y.; Hu, H. Five microRNAs in plasma as novel biomarkers for screening of early-stage non-small cell lung cancer. Respir. Res. 2014, 15, 149. [Google Scholar] [CrossRef] [PubMed]
- Yau, T.O.; Wu, C.W.; Tang, C.M.; Chen, Y.; Fang, J.; Dong, Y.; Liang, Q.; Ng, S.S.; Chan, F.K.; Sung, J.J.; et al. MicroRNA-20a in human faeces as a non-invasive biomarker for colorectal cancer. Oncotarget 2016, 7, 1559–1568. [Google Scholar] [PubMed]
- He, F.C.; Meng, W.W.; Qu, Y.H.; Zhou, M.X.; He, J.; Lv, P.; Ming, L. Expression of circulating microRNA-20a and let-7a in esophageal squamous cell carcinoma. World J. Gastroenterol. 2015, 21, 4660–4665. [Google Scholar] [PubMed]
- Conev, N.V.; Donev, I.S.; Konsoulova-Kirova, A.A.; Chervenkov, T.G.; Kashlov, J.K.; Ivanov, K.D. Serum expression levels of miR-17, miR-21, and miR-92 as potential biomarkers for recurrence after adjuvant chemotherapy in colon cancer patients. Biosci. Trends 2016, 9, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.K.; Chong, W.W.; Jin, H.; Lam, E.K.; Shin, V.Y.; Yu, J.; Poon, T.C.; Ng, S.S.; Sung, J.J. Differential expression of microRNAs in plasma of patients with colorectal cancer: A potential marker for colorectal cancer screening. Gut 2009, 58, 1375–1381. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Resnick, K.E.; Alder, H.; Hagan, J.P.; Richardson, D.L.; Croce, C.M.; Cohn, D.E. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol. Oncol. 2009, 112, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Huang, D.; Ni, S.; Peng, Z.; Sheng, W.; Du, X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int. J. Cancer 2010, 127, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, M.; Oikawa, K.; Takanashi, M.; Kudo, M.; Ohyashiki, J.; Ohyashiki, K.; Kuroda, M. Down-regulation of miR-92 in human plasma is a novel marker for acute leukemia patients. PLoS ONE 2009, 4, e5532. [Google Scholar] [CrossRef] [PubMed]
- Yuan, R.; Wang, G.; Zhi, Q.; Chen, H.; Han, Y.; Wang, B.; Kou, Z.; Hu, H.; Guo, Z.; Xue, X.; et al. Up-regulated circulating miR-106a by DNA methylation promised a potential diagnostic and prognostic marker for gastric cancer. Anticancer Agents Med. Chem. 2015. PMID:26179261. [Google Scholar]
- Koga, Y.; Yamazaki, N.; Yamamoto, Y.; Yamamoto, S.; Saito, N.; Kakugawa, Y.; Otake, Y.; Matsumoto, M.; Matsumura, Y. Fecal miR-106a is a useful marker for colorectal cancer patients with false-negative results in immunochemical fecal occult blood test. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1844–1852. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.Y.; Zhao, X.J.; Yu, Z.F.; Hu, F.L.; Liu, Y.P.; Cui, B.B.; Dong, X.S.; Zhao, Y.S. The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 7092–7101. [Google Scholar] [PubMed]
- Li, J.; Liu, Y.; Wang, C.; Deng, T.; Liang, H.; Wang, Y.; Huang, D.; Fan, Q.; Wang, X.; Ning, T.; et al. Serum miRNA expression profile as a prognostic biomarker of stage II/III colorectal adenocarcinoma. Sci. Rep. 2015, 5, 12921. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Jin, C.; Chen, W.; Xia, F.; Wang, Q.; Fan, F.; Du, J.; Guo, Y.; Lin, C.; Yang, K.; et al. Downregulation of serum miR-17 and miR-106b levels in gastric cancer and benign gastric diseases. Chin. J. Cancer Res. 2014, 26, 711–716. [Google Scholar] [PubMed]
- Jiang, L.; Li, X.; Cheng, Q.; Zhang, B.H. Plasma microRNA might as a potential biomarker for hepatocellular carcinoma and chronic liver disease screening. Tumour Biol. 2015, 36, 7167–7174. [Google Scholar] [CrossRef] [PubMed]
- Sohn, W.; Kim, J.; Kang, S.H.; Yang, S.R.; Cho, J.Y.; Cho, H.C.; Shim, S.G.; Paik, Y.H. Serum exosomal microRNAs as novel biomarkers for hepatocellular carcinoma. Exp. Mol. Med. 2015, 47, e184. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Pan, L.; Gao, J.; Ye, X.; Chen, L.; Zhang, X.; Tang, W.; Zheng, W. Prognostic value of miR-106b expression in breast cancer patients. J. Surg. Res. 2015, 195, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wang, W.; Li, F.; Zhang, H.; Liu, J. MicroRNA-106b~25 expressions in tumor tissues and plasma of patients with gastric cancers. Med. Oncol. 2014, 31, 243. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zhang, X.; Yang, Y.; Li, Z.; Du, L.; Dong, Z.; Qu, A.; Jiang, X.; Li, P.; Wang, C. Urinary cell-free microRNA-106b as a novel biomarker for detection of bladder cancer. Med. Oncol. 2014, 31, 197. [Google Scholar] [CrossRef] [PubMed]
- Ji, T.; Zheng, Z.G.; Wang, F.M.; Xu, L.J.; Li, L.F.; Cheng, Q.H.; Guo, J.F.; Ding, X.F. Differential microRNA expression by Solexa sequencing in the sera of ovarian cancer patients. Asian Pac. J. Cancer Prev. 2014, 15, 1739–1743. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Fu, R.; Yang, L.; Tu, W. miR-21 expression predicts prognosis in diffuse large B-cell lymphoma. Int. J. Clin. Exp. Pathol. 2015, 8, 15019–15024. [Google Scholar] [PubMed]
- Chen, W.; Wang, H.; Chen, H.; Liu, S.; Lu, H.; Kong, D.; Huang, X.; Kong, Q.; Lu, Z. Clinical significance and detection of microRNA-21 in serum of patients with diffuse large B-cell lymphoma in Chinese population. Eur. J. Haematol. 2014, 92, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.; Nourse, J.P.; Keane, C.; Bhatnagar, A.; Gandhi, M.K. Plasma microRNA are disease response biomarkers in classical Hodgkin lymphoma. Clin. Cancer Res. 2014, 20, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Sun, Y.; Tang, J. Serum miR-21 is a diagnostic and prognostic marker of primary central nervous system lymphoma. Neurol. Sci. 2014, 35, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Keerthana, R.; Pazhanimuthu, A.; Perumal, P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J. Biochem. Biophys. 2013, 50, 210–214. [Google Scholar] [PubMed]
- Asaga, S.; Kuo, C.; Nguyen, T.; Terpenning, M.; Giuliano, A.E.; Hoon, D.S. Direct serum assay for microRNA-21 concentrations in early and advanced breast cancer. Clin. Chem. 2011, 57, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Toraih, E.A.; Mohammed, E.A.; Farrag, S.; Ramsis, N.; Hosny, S. Pilot Study of Serum MicroRNA-21 as a Diagnostic and Prognostic Biomarker in Egyptian Breast Cancer Patients. Mol. Diagn. Ther. 2015, 19, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Usmani, A.; Shoro, A.A.; Memon, Z.; Hussain, M.; Rehman, R. Diagnostic, prognostic and predictive value of MicroRNA-21 in breast cancer patients, their daughters and healthy individuals. Am. J. Cancer Res. 2015, 5, 2484–2490. [Google Scholar] [PubMed]
- Wang, G.; Wang, L.; Sun, S.; Wu, J.; Wang, Q. Quantitative measurement of serum microRNA-21 expression in relation to breast cancer metastasis in Chinese females. Ann. Lab. Med. 2015, 35, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Matamala, N.; Vargas, M.T.; Gonzalez-Campora, R.; Minambres, R.; Arias, J.I.; Menendez, P.; Andres-Leon, E.; Gomez-Lopez, G.; Yanowsky, K.; Calvete-Candenas, J.; et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin. Chem. 2015, 61, 1098–1106. [Google Scholar] [CrossRef] [PubMed]
- Erbes, T.; Hirschfeld, M.; Rucker, G.; Jaeger, M.; Boas, J.; Iborra, S.; Mayer, S.; Gitsch, G.; Stickeler, E. Feasibility of urinary microRNA detection in breast cancer patients and its potential as an innovative non-invasive biomarker. BMC Cancer 2015, 15, 193. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.J.; Gu, R.M.; Zhu, M.; Wen, X.; Li, J.T.; Zhang, Y.Y.; Zhang, X.M.; Chen, S.Q. Plasma post-operative miR-21 expression in the prognosis of gastric cancers. Asian Pac. J. Cancer Prev. 2013, 14, 7551–7554. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Bai, Z.; Zhang, J.; Meng, H.; Cai, J.; Deng, W.; Bi, J.; Ma, X.; Zhang, Z. Serum microRNA-21 levels are related to tumor size in gastric cancer patients but cannot predict prognosis. Oncol. Lett. 2013, 6, 1733–1737. [Google Scholar] [PubMed]
- Kim, S.Y.; Jeon, T.Y.; Choi, C.I.; Kim, D.H.; Kim, D.H.; Kim, G.H.; Ryu, D.Y.; Lee, B.E.; Kim, H.H. Validation of circulating miRNA biomarkers for predicting lymph node metastasis in gastric cancer. J. Mol. Diagn. 2013, 15, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Shiotani, A.; Murao, T.; Kimura, Y.; Matsumoto, H.; Kamada, T.; Kusunoki, H.; Inoue, K.; Uedo, N.; Iishi, H.; Haruma, K. Identification of serum miRNAs as novel non-invasive biomarkers for detection of high risk for early gastric cancer. Br. J. Cancer 2013, 109, 2323–2330. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Tsujiura, M.; Konishi, H.; Takeshita, H.; Nagata, H.; Kawaguchi, T.; Hirajima, S.; Arita, T.; Shiozaki, A.; et al. Prognostic impact of circulating miR-21 in the plasma of patients with gastric carcinoma. Anticancer Res. 2013, 33, 271–276. [Google Scholar] [PubMed]
- Wu, J.; Li, G.; Wang, Z.; Yao, Y.; Chen, R.; Pu, X.; Wang, J. Circulating MicroRNA-21 Is a Potential Diagnostic Biomarker in Gastric Cancer. Dis. Markers 2015, 2015, 435656. [Google Scholar] [CrossRef] [PubMed]
- Ilhan-Mutlu, A.; Wagner, L.; Wohrer, A.; Furtner, J.; Widhalm, G.; Marosi, C.; Preusser, M. Plasma MicroRNA-21 concentration may be a useful biomarker in glioblastoma patients. Cancer Investig. 2012, 30, 615–621. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Z.; Xi, Q.H.; Ge, W.L.; Zhang, X.Q. Identification of serum microRNA-21 as a biomarker for early detection and prognosis in human epithelial ovarian cancer. Asian Pac. J. Cancer Prev. 2013, 14, 1057–1060. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, J.; Chang, P.; LeBlanc, A.; Li, D.; Abbruzzesse, J.L.; Frazier, M.L.; Killary, A.M.; Sen, S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. (Phila) 2009, 2, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Abue, M.; Yokoyama, M.; Shibuya, R.; Tamai, K.; Yamaguchi, K.; Sato, I.; Tanaka, N.; Hamada, S.; Shimosegawa, T.; Sugamura, K.; et al. Circulating miR-483-3p and miR-21 is highly expressed in plasma of pancreatic cancer. Int. J. Oncol. 2015, 46, 539–547. [Google Scholar] [CrossRef] [PubMed]
- Ali, S.; Dubaybo, H.; Brand, R.E.; Sarkar, F.H. Differential Expression of MicroRNAs in Tissues and Plasma Co-exists as a Biomarker for Pancreatic Cancer. J. Cancer Sci. Ther. 2015, 7, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Sun, Y.W.; Liu, D.J.; Zhang, J.F.; Li, J.; Hua, R. MicroRNAs in stool samples as potential screening biomarkers for pancreatic ductal adenocarcinoma cancer. Am. J. Cancer Res. 2014, 4, 663–673. [Google Scholar] [PubMed]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.; Tang, W.; Yuan, W.; Yu, Q.; Zuo, W.; Xu, C.; Ma, J. Expression and clinical significance of plasma small RNA in patients with pancreatic cancer. Zhonghua Zhong Liu Za Zhi 2014, 36, 351–354. [Google Scholar] [PubMed]
- Humeau, M.; Vignolle-Vidoni, A.; Sicard, F.; Martins, F.; Bournet, B.; Buscail, L.; Torrisani, J.; Cordelier, P. Salivary MicroRNA in Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0130996. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.L.; Yang, L.F.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Zhu, Y.P.; Shen, Y.J.; Shi, G.H.; Ye, D.W. Serum miRNA-21: Elevated levels in patients with metastatic hormone-refractory prostate cancer and potential predictive factor for the efficacy of docetaxel-based chemotherapy. Prostate 2011, 71, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Basati, G.; Emami Razavi, A.; Abdi, S.; Mirzaei, A. Elevated level of microRNA-21 in the serum of patients with colorectal cancer. Med. Oncol. 2014, 31, 205. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Takahashi, M.; Hur, K.; Nagasaka, T.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J. Natl. Cancer Inst. 2013, 105, 849–859. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Zhou, Z.G.; Chen, R.; Wang, M.J.; Zhou, B.; Li, Y.; Sun, X.F. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumour Biol. 2013, 34, 2175–2181. [Google Scholar] [CrossRef] [PubMed]
- Menendez, P.; Padilla, D.; Villarejo, P.; Palomino, T.; Nieto, P.; Menendez, J.M.; Rodriguez-Montes, J.A. Prognostic implications of serum microRNA-21 in colorectal cancer. J. Surg. Oncol. 2013, 108, 369–373. [Google Scholar] [CrossRef] [PubMed]
- Beckett, E.L.; Martin, C.; Choi, J.H.; King, K.; Niblett, S.; Boyd, L.; Duesing, K.; Yates, Z.; Veysey, M.; Lucock, M. Folate status, folate-related genes and serum miR-21 expression: Implications for miR-21 as a biomarker. BBA Clin. 2015, 4, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.S.; Li, B.J.; Lu, H.W.; Chen, Y.; Lu, C.; Zhu, R.X.; Liu, S.H.; Yi, Q.T.; Li, J.; Song, C.H. Serum miR-152, miR-148a, miR-148b, and miR-21 as novel biomarkers in non-small cell lung cancer screening. Tumour Biol. 2015, 36, 3035–3042. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.O.; Gazala, S.; Razzak, R.; Guo, L.; Ghosh, S.; Roa, W.H.; Bedard, E.L. Non-small cell lung cancer detection using microRNA expression profiling of bronchoalveolar lavage fluid and sputum. Anticancer Res. 2015, 35, 1873–1880. [Google Scholar] [PubMed]
- Zhao, W.; Zhao, J.J.; Zhang, L.; Xu, Q.F.; Zhao, Y.M.; Shi, X.Y.; Xu, A.G. Serum miR-21 level: A potential diagnostic and prognostic biomarker for non-small cell lung cancer. Int. J. Clin. Exp. Med. 2015, 8, 14759–14763. [Google Scholar] [PubMed]
- Hsu, C.M.; Lin, P.M.; Wang, Y.M.; Chen, Z.J.; Lin, S.F.; Yang, M.Y. Circulating miRNA is a novel marker for head and neck squamous cell carcinoma. Tumour Biol. 2012, 33, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Morimura, R.; Nagata, H.; Kosuga, T.; Iitaka, D.; Konishi, H.; Shiozaki, A.; et al. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. Br. J. Cancer 2011, 105, 104–111. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Chen, G.; Zhang, X.; Li, D.; Huang, J.; Yang, C.; Zhang, P.; Qin, Y.; Duan, Y.; Gong, B.; et al. Salivary microRNAs as promising biomarkers for detection of esophageal cancer. PLoS ONE 2013, 8, e57502. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, C.; Jiang, W.; Huang, D.; Xu, L.; Yang, Q.; Zheng, L.; Wang, X.; Hu, L. Serum miR-21, miR-26a and miR-101 as potential biomarkers of hepatocellular carcinoma. Clin. Res. Hepatol. Gastroenterol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Bai, Z.; Song, J.; Yang, Y.; Wang, J.; Han, W.; Zhang, J.; Meng, H.; Ma, X.; Yang, Y.; et al. Differential expression of serum miR-126, miR-141 and miR-21 as novel biomarkers for early detection of liver metastasis in colorectal cancer. Chin. J. Cancer Res. 2014, 26, 95–103. [Google Scholar] [PubMed]
- Wang, H.; Hou, L.; Li, A.; Duan, Y.; Gao, H.; Song, X. Expression of serum exosomal microRNA-21 in human hepatocellular carcinoma. BioMed Res. Int. 2014, 2014, 864894. [Google Scholar] [CrossRef] [PubMed]
- Amr, K.S.; Ezzat, W.M.; Elhosary, Y.A.; Hegazy, A.E.; Fahim, H.H.; Kamel, R.R. The potential role of miRNAs 21 and 199-a in early diagnosis of hepatocellular carcinoma. Gene 2016, 575, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, T.; Eguchi, H.; Nagano, H.; Kobayashi, S.; Akita, H.; Hama, N.; Wada, H.; Kawamoto, K.; Tomokuni, A.; Tomimaru, Y.; et al. Plasma miR-21 is a novel diagnostic biomarker for biliary tract cancer. Cancer Sci. 2013, 104, 1626–1631. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Luo, H.N.; Tian, W.D.; Lu, J.; Li, G.; Wang, L.; Zhang, B.; Liang, B.J.; Peng, X.H.; Lin, S.X.; et al. Diagnostic and prognostic value of plasma microRNA deregulation in nasopharyngeal carcinoma. Cancer Biol. Ther. 2013, 14, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, L.; Liu, P.; Yang, S.; Ye, S.; Xu, W.; Liu, X. A three-plasma miRNA signature serves as novel biomarkers for osteosarcoma. Med. Oncol. 2013, 30, 340. [Google Scholar] [CrossRef] [PubMed]
- Hong, Q.; Fang, J.; Pang, Y.; Zheng, J. Prognostic value of the microRNA-29 family in patients with primary osteosarcomas. Med. Oncol. 2014, 31, 37. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.G.; Gu, J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012, 36, e61–e67. [Google Scholar] [CrossRef] [PubMed]
- Brunet Vega, A.; Pericay, C.; Moya, I.; Ferrer, A.; Dotor, E.; Pisa, A.; Casalots, A.; Serra-Aracil, X.; Oliva, J.C.; Ruiz, A.; et al. microRNA expression profile in stage III colorectal cancer: Circulating miR-18a and miR-29a as promising biomarkers. Oncol. Rep. 2013, 30, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 2012, 413, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Lu, Z.; Li, H.; Lu, J.; Guo, L.; Ge, Q. Next-generation sequencing of microRNAs for breast cancer detection. J. Biomed. Biotechnol. 2011, 2011, 597145. [Google Scholar] [CrossRef] [PubMed]
- Basati, G.; Razavi, A.E.; Pakzad, I.; Malayeri, F.A. Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Yang, I.P.; Tsai, H.L.; Huang, C.W.; Huang, M.Y.; Hou, M.F.; Juo, S.H.; Wang, J.Y. The functional significance of microRNA-29c in patients with colorectal cancer: A potential circulating biomarker for predicting early relapse. PLoS ONE 2013, 8, e66842. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; He, J.; Chen, D.; Zhang, B.; Xu, L.; Ma, H.; Liu, X.; Zhang, Y.; Le, H. Expression of miR-29c, miR-93, and miR-429 as potential biomarkers for detection of early stage non-small lung cancer. PLoS ONE 2014, 9, e87780. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Xiang, J.; Wu, M.; Xiong, W.; Tang, H.; Deng, M.; Li, X.; Liao, Q.; Su, B.; Luo, Z.; et al. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS ONE 2012, 7, e46367. [Google Scholar] [CrossRef] [PubMed]
- Cazzoli, R.; Buttitta, F.; di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. MicroRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Chiam, K.; Wang, T.; Watson, D.I.; Mayne, G.C.; Irvine, T.S.; Bright, T.; Smith, L.; White, I.A.; Bowen, J.M.; Keefe, D.; et al. Circulating Serum Exosomal miRNAs As Potential Biomarkers for Esophageal Adenocarcinoma. J. Gastrointest. Surg. 2015, 19, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.H.; Zhang, G.L.; Li, H.R.; Luo, J.D.; Li, Z.X.; Chen, G.M.; Yang, J. A panel of five circulating microRNAs as potential biomarkers for prostate cancer. Prostate 2012, 72, 1443–1452. [Google Scholar] [CrossRef] [PubMed]
- Kachakova, D.; Mitkova, A.; Popov, E.; Popov, I.; Vlahova, A.; Dikov, T.; Christova, S.; Mitev, V.; Slavov, C.; Kaneva, R. Combinations of serum prostate-specific antigen and plasma expression levels of let-7c, miR-30c, miR-141, and miR-375 as potential better diagnostic biomarkers for prostate cancer. DNA Cell Biol. 2015, 34, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Rani, S.; Gately, K.; Crown, J.; O’Byrne, K.; O’Driscoll, L. Global analysis of serum microRNAs as potential biomarkers for lung adenocarcinoma. Cancer Biol. Ther. 2013, 14, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Le, H.B.; Zhu, W.Y.; Chen, D.D.; He, J.Y.; Huang, Y.Y.; Liu, X.G.; Zhang, Y.K. Evaluation of dynamic change of serum miR-21 and miR-24 in pre- and post-operative lung carcinoma patients. Med. Oncol. 2012, 29, 3190–3197. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chen, X.; Zhao, Y.; Tian, T.; Jin, G.; Shu, Y.; Chen, Y.; Xu, L.; Zen, K.; Zhang, C.; et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010, 28, 1721–1726. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, S.; Steele, R.; Shrivastava, S.; Chakraborty, S.; di Bisceglie, A.M.; Ray, R.B. Serum miR-30e and miR-223 as Novel Noninvasive Biomarkers for Hepatocellular Carcinoma. Am. J. Pathol. 2016, 186, 242–247. [Google Scholar] [CrossRef] [PubMed]
- Franchina, T.; Amodeo, V.; Bronte, G.; Savio, G.; Ricciardi, G.R.; Picciotto, M.; Russo, A.; Giordano, A.; Adamo, V. Circulating miR-22, miR-24 and miR-34a as novel predictive biomarkers to pemetrexed-based chemotherapy in advanced non-small cell lung cancer. J. Cell. Physiol. 2014, 229, 97–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; Jia, J.; Ling, S.; Liu, Y.; Yang, S.; Shao, Z. A causal role for circulating miR-34b in osteosarcoma. Eur. J. Surg. Oncol. 2014, 40, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Feng, J.; Tang, L.; Liao, L.; Xu, Q.; Zhu, S. The regulation and function of miR-21-FOXO3a-miR-34b/c signaling in breast cancer. Int. J. Mol. Sci. 2015, 16, 3148–3162. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Wang, X.; Li, J.; Gu, K.; Lv, L.; Zhang, S.; Che, D.; Cao, J.; Jin, S.; Yu, Y. MiR-34c-3p functions as a tumour suppressor by inhibiting eIF4E expression in non-small cell lung cancer. Cell Prolif. 2015, 48, 582–592. [Google Scholar] [CrossRef] [PubMed]
- Park, N.J.; Zhou, H.; Elashoff, D.; Henson, B.S.; Kastratovic, D.A.; Abemayor, E.; Wong, D.T. Salivary microRNA: Discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009, 15, 5473–5477. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.J.; Zheng, Y.H.; Wang, P.; Zhang, J.Z. Serum miR-125a-5p, miR-145 and miR-146a as diagnostic biomarkers in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 2015, 8, 765–771. [Google Scholar] [PubMed]
- Hsieh, T.H.; Hsu, C.Y.; Tsai, C.F.; Long, C.Y.; Chai, C.Y.; Hou, M.F.; Lee, J.N.; Wu, D.C.; Wang, S.C.; Tsai, E.M. miR-125a-5p is a prognostic biomarker that targets HDAC4 to suppress breast tumorigenesis. Oncotarget 2015, 6, 494–509. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Zhou, Z.; Xu, Z.; Li, G.; Dong, P.; Chen, Z.; Lin, D.; Chen, B.; Yu, F. Serum microRNA-125a-5p, a useful biomarker in liver diseases, correlates with disease progression. Mol. Med. Rep. 2015, 12, 1584–1590. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tan, G.; Dong, L.; Cheng, L.; Li, K.; Wang, Z.; Luo, H. Circulating miR-125b as a marker predicting chemoresistance in breast cancer. PLoS ONE 2012, 7, e34210. [Google Scholar] [CrossRef] [PubMed]
- Yuxia, M.; Zhennan, T.; Wei, Z. Circulating miR-125b is a novel biomarker for screening non-small-cell lung cancer and predicts poor prognosis. J. Cancer Res. Clin. Oncol. 2012, 138, 2045–2050. [Google Scholar] [CrossRef] [PubMed]
- Cui, E.H.; Li, H.J.; Hua, F.; Wang, B.; Mao, W.; Feng, X.R.; Li, J.Y.; Wang, X. Serum microRNA 125b as a diagnostic or prognostic biomarker for advanced NSCLC patients receiving cisplatin-based chemotherapy. Acta Pharmacol. Sin. 2013, 34, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.L.; Ye, D.X.; Wu, J.J. Expression and clinical significance of plasma microRNA-125b level in patients with oral squamous cell carcinoma. Shanghai Kou Qiang Yi Xue 2015, 24, 71–75. [Google Scholar] [PubMed]
- Yamada, A.; Horimatsu, T.; Okugawa, Y.; Nishida, N.; Honjo, H.; Ida, H.; Kou, T.; Kusaka, T.; Sasaki, Y.; Yagi, M.; et al. Serum miR-21, miR-29a, and miR-125b Are Promising Biomarkers for the Early Detection of Colorectal Neoplasia. Clin. Cancer Res. 2015, 21, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Chen, D.; Lv, T.; Li, G.; Qu, S. Serum MicroRNA-125b as a Potential Biomarker for Glioma Diagnosis. Mol. Neurobiol. 2016, 53, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Alegre, E.; Sanmamed, M.F.; Rodriguez, C.; Carranza, O.; Martin-Algarra, S.; Gonzalez, A. Study of circulating microRNA-125b levels in serum exosomes in advanced melanoma. Arch. Pathol. Lab. Med. 2014, 138, 828–832. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Hou, J.; Jin, W.; Li, J.; Yue, Y.; Jin, H.; Wang, X. Increased circulating microRNA-155 as a potential biomarker for breast cancer screening: A meta-analysis. Molecules 2014, 19, 6282–6293. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Mao, Q.; Liu, Y.; Hao, X.; Zhang, S.; Zhang, J. Analysis of miR-205 and miR-155 expression in the blood of breast cancer patients. Chin. J. Cancer Res. 2013, 25, 46–54. [Google Scholar] [PubMed]
- Sun, Y.; Wang, M.; Lin, G.; Sun, S.; Li, X.; Qi, J.; Li, J. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS ONE 2012, 7, e47003. [Google Scholar] [CrossRef] [PubMed]
- Eichelser, C.; Flesch-Janys, D.; Chang-Claude, J.; Pantel, K.; Schwarzenbach, H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin. Chem. 2013, 59, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.C.; Fan, Y.S.; Chen, H.B.; Zhao, D.W. Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol. 2015, 36, 1619–1625. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Chang, J.; Wang, H.; Zhang, G. Potential diagnostic value of miR-155 in serum from lung adenocarcinoma patients. Oncol. Rep. 2014, 31, 351–357. [Google Scholar] [PubMed]
- Liu, R.; Liao, J.; Yang, M.; Shi, Y.; Peng, Y.; Wang, Y.; Pan, E.; Guo, W.; Pu, Y.; Yin, L. Circulating miR-155 expression in plasma: A potential biomarker for early diagnosis of esophageal cancer in humans. J. Toxicol. Environ. Health A 2012, 75, 1154–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhi, F.; Cao, X.; Xie, X.; Wang, B.; Dong, W.; Gu, W.; Ling, Y.; Wang, R.; Yang, Y.; Liu, Y. Identification of circulating microRNAs as potential biomarkers for detecting acute myeloid leukemia. PLoS ONE 2013, 8, e56718. [Google Scholar] [CrossRef] [PubMed]
- Fang, C.; Zhu, D.X.; Dong, H.J.; Zhou, Z.J.; Wang, Y.H.; Liu, L.; Fan, L.; Miao, K.R.; Liu, P.; Xu, W.; et al. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann. Hematol. 2012, 91, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Ferrajoli, A.; Shanafelt, T.D.; Ivan, C.; Shimizu, M.; Rabe, K.G.; Nouraee, N.; Ikuo, M.; Ghosh, A.K.; Lerner, S.; Rassenti, L.Z.; et al. Prognostic value of miR-155 in individuals with monoclonal B-cell lymphocytosis and patients with B chronic lymphocytic leukemia. Blood 2013, 122, 1891–1899. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, K.; Sasaki, D.; Tsuruda, K.; Inokuchi, N.; Nagai, K.; Hasegawa, H.; Yanagihara, K.; Kamihira, S. Impact of miR-155 and miR-126 as novel biomarkers on the assessment of disease progression and prognosis in adult T-cell leukemia. Cancer Epidemiol. 2012, 36, 560–565. [Google Scholar] [CrossRef] [PubMed]
- Zuberi, M.; Mir, R.; Das, J.; Ahmad, I.; Javid, J.; Yadav, P.; Masroor, M.; Ahmad, S.; Ray, P.C.; Saxena, A. Expression of serum miR-200a, miR-200b, and miR-200c as candidate biomarkers in epithelial ovarian cancer and their association with clinicopathological features. Clin. Transl. Oncol. 2015, 17, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Kan, C.W.; Hahn, M.A.; Gard, G.B.; Maidens, J.; Huh, J.Y.; Marsh, D.J.; Howell, V.M. Elevated levels of circulating microRNA-200 family members correlate with serous epithelial ovarian cancer. BMC Cancer 2012, 12, 627. [Google Scholar] [CrossRef] [PubMed]
- Antolin, S.; Calvo, L.; Blanco-Calvo, M.; Santiago, M.P.; Lorenzo-Patino, M.J.; Haz-Conde, M.; Santamarina, I.; Figueroa, A.; Anton-Aparicio, L.M.; Valladares-Ayerbes, M. Circulating miR-200c and miR-141 and outcomes in patients with breast cancer. BMC Cancer 2015, 15, 297. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.C.; Wu, J. MicroRNA-200c and microRNA-141 as potential diagnostic and prognostic biomarkers for ovarian cancer. Tumour Biol. 2015, 36, 4843–4850. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.M.; Castillo, L.; Mahon, K.L.; Chiam, K.; Lee, B.Y.; Nguyen, Q.; Boyer, M.J.; Stockler, M.R.; Pavlakis, N.; Marx, G.; et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br. J. Cancer 2014, 110, 2462–2471. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Zhou, T.; Liu, Z.L.; Tian, H.P.; Xia, S.S. Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Mol. Clin. Oncol. 2013, 1, 379–384. [Google Scholar] [PubMed]
- Toiyama, Y.; Hur, K.; Tanaka, K.; Inoue, Y.; Kusunoki, M.; Boland, C.R.; Goel, A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann. Surg. 2014, 259, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.P.; Sun, F.B.; Li, S.J. Serum miR-200c expression level as a prognostic biomarker for gastric cancer. Genet. Mol. Res. 2015, 14, 15913–15920. [Google Scholar] [CrossRef] [PubMed]
- Valladares-Ayerbes, M.; Reboredo, M.; Medina-Villaamil, V.; Iglesias-Diaz, P.; Lorenzo-Patino, M.J.; Haz, M.; Santamarina, I.; Blanco, M.; Fernandez-Tajes, J.; Quindos, M.; et al. Circulating miR-200c as a diagnostic and prognostic biomarker for gastric cancer. J. Transl. Med. 2012, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.G.; Zhu, W.Y.; Huang, Y.Y.; Ma, L.N.; Zhou, S.Q.; Wang, Y.K.; Zeng, F.; Zhou, J.H.; Zhang, Y.K. High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med. Oncol. 2012, 29, 618–626. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Miyata, H.; Yamasaki, M.; Sugimura, K.; Takahashi, T.; Kurokawa, Y.; Nakajima, K.; Takiguchi, S.; Mori, M.; Doki, Y. Circulating miR-200c levels significantly predict response to chemotherapy and prognosis of patients undergoing neoadjuvant chemotherapy for esophageal cancer. Ann. Surg. Oncol. 2013, 20, S607–S615. [Google Scholar] [CrossRef] [PubMed]
- Kriebel, S.; Schmidt, D.; Holdenrieder, S.; Goltz, D.; Kristiansen, G.; Moritz, R.; Fisang, C.; Muller, S.C.; Ellinger, J. Analysis of tissue and serum microRNA expression in patients with upper urinary tract urothelial cancer. PLoS ONE 2015, 10, e0117284. [Google Scholar] [CrossRef] [PubMed]
- Brase, J.C.; Johannes, M.; Schlomm, T.; Falth, M.; Haese, A.; Steuber, T.; Beissbarth, T.; Kuner, R.; Sultmann, H. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int. J. Cancer 2011, 128, 608–616. [Google Scholar] [CrossRef] [PubMed]
- Bryant, R.J.; Pawlowski, T.; Catto, J.W.; Marsden, G.; Vessella, R.L.; Rhees, B.; Kuslich, C.; Visakorpi, T.; Hamdy, F.C. Changes in circulating microRNA levels associated with prostate cancer. Br. J. Cancer. 2012, 106, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ma, Y.Y.; Wang, J.; Zeng, X.F.; Li, R.; Kang, W.; Hao, X.K. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Oncol. Targets Ther. 2016, 9, 139–148. [Google Scholar]
- Zhang, H.L.; Qin, X.J.; Cao, D.L.; Zhu, Y.; Yao, X.D.; Zhang, S.L.; Dai, B.; Ye, D.W. An elevated serum miR-141 level in patients with bone-metastatic prostate cancer is correlated with more bone lesions. Asian J. Androl. 2013, 15, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Zhang, L.; Cogdell, D.E.; Zheng, H.; Schetter, A.J.; Nykter, M.; Harris, C.C.; Chen, K.; Hamilton, S.R.; Zhang, W. Circulating plasma miR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS ONE 2011, 6, e17745. [Google Scholar] [CrossRef] [PubMed]
- Ho, A.S.; Huang, X.; Cao, H.; Christman-Skieller, C.; Bennewith, K.; Le, Q.T.; Koong, A.C. Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer. Transl. Oncol. 2010, 3, 109–113. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Li, G.; Peoc’h, M.; Genin, C.; Gigante, M. Serum miR-210 as a novel biomarker for molecular diagnosis of clear cell renal cell carcinoma. Exp. Mol. Pathol. 2013, 94, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, H.; Kanda, Y.; Sejima, T.; Osaki, M.; Okada, F.; Takenaka, A. Serum miR-210 as a potential biomarker of early clear cell renal cell carcinoma. Int. J. Oncol. 2014, 44, 53–58. [Google Scholar] [PubMed]
- Jung, E.J.; Santarpia, L.; Kim, J.; Esteva, F.J.; Moretti, E.; Buzdar, A.U.; di Leo, A.; Le, X.F.; Bast, R.C., Jr.; Park, S.T.; et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer 2012, 118, 2603–2614. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Raimondo, M.; Guha, S.; Chen, J.; Diao, L.; Dong, X.; Wallace, M.B.; Killary, A.M.; Frazier, M.L.; Woodward, T.A.; et al. Circulating microRNAs in Pancreatic Juice as Candidate Biomarkers of Pancreatic Cancer. J. Cancer 2014, 5, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Qu, A.; Liu, J.; Wang, R.; Liu, Y.; Li, G.; Duan, W.; Fang, Q.; Jiang, X.; Wang, L.; et al. Serum miR-210 Contributes to Tumor Detection, Stage Prediction and Dynamic Surveillance in Patients with Bladder Cancer. PLoS ONE 2015, 10, e0135168. [Google Scholar] [CrossRef] [PubMed]
- Eissa, S.; Matboli, M.; Essawy, N.O.; Kotb, Y.M. Integrative functional genetic-epigenetic approach for selecting genes as urine biomarkers for bladder cancer diagnosis. Tumour Biol. 2015, 36, 9545–9552. [Google Scholar] [CrossRef] [PubMed]
- Lai, N.S.; Wu, D.G.; Fang, X.G.; Lin, Y.C.; Chen, S.S.; Li, Z.B.; Xu, S.S. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br. J. Cancer 2015, 112, 1241–1246. [Google Scholar] [CrossRef] [PubMed]
- Zhan, M.; Li, Y.; Hu, B.; He, X.; Huang, J.; Zhao, Y.; Fu, S.; Lu, L. Serum microRNA-210 as a predictive biomarker for treatment response and prognosis in patients with hepatocellular carcinoma undergoing transarterial chemoembolization. J. Vasc. Int. Radiol. 2014, 25, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Pu, X.X.; Huang, G.L.; Guo, H.Q.; Guo, C.C.; Li, H.; Ye, S.; Ling, S.; Jiang, L.; Tian, Y.; Lin, T.Y. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J. Gastroenterol. Hepatol. 2010, 25, 1674–1680. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.Q.; Huang, G.L.; Guo, C.C.; Pu, X.X.; Lin, T.Y. Diagnostic and prognostic value of circulating miR-221 for extranodal natural killer/T-cell lymphoma. Dis. Markers 2010, 29, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, S.S.; Guzel, E.; Karatas, O.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. MiR-221 as a pre- and postoperative plasma biomarker for larynx cancer patients. Laryngoscope 2015, 125, E377–E381. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Pang, B.; Xin, T.; Guo, H.; Xing, Y.; Xu, S.; Feng, B.; Liu, B.; Pang, Q. Plasma miR-221/222 Family as Novel Descriptive and Prognostic Biomarkers for Glioma. Mol. Neurobiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; He, Q.Y.; Luo, C.Q.; Qian, L.Y. Circulating miR-221 expression level and prognosis of cutaneous malignant melanoma. Med. Sci. Monit. 2014, 20, 2472–2477. [Google Scholar] [PubMed]
- Teixeira, A.L.; Ferreira, M.; Silva, J.; Gomes, M.; Dias, F.; Santos, J.I.; Mauricio, J.; Lobo, F.; Medeiros, R. Higher circulating expression levels of miR-221 associated with poor overall survival in renal cell carcinoma patients. Tumour Biol. 2014, 35, 4057–4066. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Somlo, G.; Yu, Y.; Palomares, M.R.; Li, A.X.; Zhou, W.; Chow, A.; Yen, Y.; Rossi, J.J.; Gao, H.; et al. De novo sequencing of circulating miRNAs identifies novel markers predicting clinical outcome of locally advanced breast cancer. J. Transl. Med. 2012, 10, 42. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Jiang, L.; Sun, C.; Guo, L.; Lin, M.; Huang, J.; Zhu, L. Decreased circulating miR-375: A potential biomarker for patients with non-small-cell lung cancer. Gene 2014, 534, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.C.; Xie, W.; Yang, M.; Hsieh, C.L.; Drouin, S.; Lee, G.S.; Kantoff, P.W. Expression differences of circulating microRNAs in metastatic castration resistant prostate cancer and low-risk, localized prostate cancer. Prostate 2013, 73, 346–354. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Li, M.; Hu, C.; Duan, H. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol. Biol. Rep. 2014, 41, 1257–1266. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hou, P.; Wu, Z.; Wang, T.; Nie, Y. Circulating miR-375 and miR-199a-3p as potential biomarkers for the diagnosis of hepatocellular carcinoma. Tumour Biol. 2015, 36, 4501–4507. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Li, M.; Wang, M.; Yan, D.; Feng, G.; An, G. The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer 2014, 14, 714. [Google Scholar] [CrossRef] [PubMed]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Boyerinas, B.; Park, S.M.; Hau, A.; Murmann, A.E.; Peter, M.E. The role of let-7 in cell differentiation and cancer. Endocr. Relat. Cancer 2010, 17, F19–F36. [Google Scholar] [CrossRef] [PubMed]
- Johnson, S.M.; Grosshans, H.; Shingara, J.; Byrom, M.; Jarvis, R.; Cheng, A.; Labourier, E.; Reinert, K.L.; Brown, D.; Slack, F.J. RAS is regulated by the let-7 microRNA family. Cell 2005, 120, 635–647. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.S.; Erkeland, S.J.; Pester, R.E.; Chen, C.Y.; Ebert, M.S.; Sharp, P.A.; Jacks, T. Suppression of non-small cell lung tumor development by the let-7 microRNA family. Proc. Natl. Acad. Sci. USA 2008, 105, 3903–3908. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Hemann, M.T.; Bartel, D.P. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science 2007, 315, 1576–1579. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lu, Y.; Toh, S.T.; Sung, W.K.; Tan, P.; Chow, P.; Chung, A.Y.; Jooi, L.L.; Lee, C.G. Lethal-7 is down-regulated by the hepatitis B virus x protein and targets signal transducer and activator of transcription 3. J. Hepatol. 2010, 53, 57–66. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Duan, C.; Chen, J.; Ou-Yang, X.; Zhang, Z.; Li, C.; Peng, H. Let-7a elevates p21(WAF1) levels by targeting of NIRF and suppresses the growth of A549 lung cancer cells. FEBS Lett. 2009, 583, 3501–3507. [Google Scholar] [CrossRef] [PubMed]
- Sampson, V.B.; Rong, N.H.; Han, J.; Yang, Q.; Aris, V.; Soteropoulos, P.; Petrelli, N.J.; Dunn, S.P.; Krueger, L.J. MicroRNA let-7a down-regulates MYC and reverts MYC-induced growth in Burkitt lymphoma cells. Cancer Res. 2007, 67, 9762–9770. [Google Scholar] [CrossRef] [PubMed]
- Akao, Y.; Nakagawa, Y.; Naoe, T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol. Pharm. Bull. 2006, 29, 903–906. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef] [PubMed]
- Legesse-Miller, A.; Elemento, O.; Pfau, S.J.; Forman, J.J.; Tavazoie, S.; Coller, H.A. Let-7 Overexpression leads to an increased fraction of cells in G2/M, direct down-regulation of Cdc34, and stabilization of Wee1 kinase in primary fibroblasts. J. Biol. Chem. 2009, 284, 6605–6609. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar] [CrossRef] [PubMed]
- Schultz, J.; Lorenz, P.; Gross, G.; Ibrahim, S.; Kunz, M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008, 18, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Tsang, W.P.; Kwok, T.T. Let-7a microRNA suppresses therapeutics-induced cancer cell death by targeting caspase-3. Apoptosis 2008, 13, 1215–1222. [Google Scholar] [CrossRef] [PubMed]
- Tokumaru, S.; Suzuki, M.; Yamada, H.; Nagino, M.; Takahashi, T. Let-7 regulates Dicer expression and constitutes a negative feedback loop. Carcinogenesis 2008, 29, 2073–2077. [Google Scholar] [CrossRef] [PubMed]
- Forman, J.J.; Legesse-Miller, A.; Coller, H.A. A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc. Natl. Acad. Sci. USA 2008, 105, 14879–14884. [Google Scholar] [CrossRef] [PubMed]
- Cookson, V.J.; Bentley, M.A.; Hogan, B.V.; Horgan, K.; Hayward, B.E.; Hazelwood, L.D.; Hughes, T.A. Circulating microRNA profiles reflect the presence of breast tumours but not the profiles of microRNAs within the tumours. Cell. Oncol. 2012, 35, 301–308. [Google Scholar] [CrossRef] [PubMed]
- Fayyad-Kazan, H.; Bitar, N.; Najar, M.; Lewalle, P.; Fayyad-Kazan, M.; Badran, R.; Hamade, E.; Daher, A.; Hussein, N.; ElDirani, R.; et al. Circulating miR-150 and miR-342 in plasma are novel potential biomarkers for acute myeloid leukemia. J. Transl. Med. 2013, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Joosse, S.A.; Muller, V.; Steinbach, B.; Pantel, K.; Schwarzenbach, H. Circulating cell-free cancer-testis MAGE-A RNA, BORIS RNA, let-7b and miR-202 in the blood of patients with breast cancer and benign breast diseases. Br. J. Cancer 2014, 111, 909–917. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Yan, W.; Wang, Y.; Sun, G.; Luo, H.; Zhang, J.; Wang, X.; You, Y.; Yang, Z.; Liu, N. MicroRNA-10b induces glioma cell invasion by modulating MMP-14 and uPAR expression via HOXD10. Brain Res. 2011, 1389, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Luo, A.; Cai, Y.; Su, Q.; Ding, F.; Chen, H.; Liu, Z. MicroRNA-10b promotes migration and invasion through KLF4 in human esophageal cancer cell lines. J. Biol. Chem. 2010, 285, 7986–7994. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, S.A.; Yip, G.W.; Stock, C.; Pan, J.W.; Neubauer, C.; Poeter, M.; Pupjalis, D.; Koo, C.Y.; Kelsch, R.; Schule, R.; et al. Targeting of syndecan-1 by microRNA miR-10b promotes breast cancer cell motility and invasiveness via a Rho-GTPase- and E-cadherin-dependent mechanism. Int. J. Cancer 2012, 131, E884–E896. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.G.; Kong, L.M.; Zhou, P.; Yang, X.L.; Huang, J.G.; Zhang, H.L.; Lu, N. miR-10b is overexpressed in hepatocellular carcinoma and promotes cell proliferation, migration and invasion through RhoC, uPAR and MMPs. J. Transl. Med. 2014, 12, 234. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhu, J.; Cao, H.; Ren, H.; Fang, X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int. J. Oncol. 2012, 40, 1553–1560. [Google Scholar] [PubMed]
- Ma, Z.; Chen, Y.; Min, L.; Li, L.; Huang, H.; Li, J.; Yan, Q.; Song, P.; Dai, L.; Yao, X. Augmented miR-10b expression associated with depressed expression of its target gene KLF4 involved in gastric carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 5071–5079. [Google Scholar] [PubMed]
- Tsukerman, P.; Stern-Ginossar, N.; Gur, C.; Glasner, A.; Nachmani, D.; Bauman, Y.; Yamin, R.; Vitenshtein, A.; Stanietsky, N.; Bar-Mag, T.; et al. miR-10b downregulates the stress-induced cell surface molecule MICB, a critical ligand for cancer cell recognition by natural killer cells. Cancer Res. 2012, 72, 5463–5472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, J.; Wang, B.; Ren, J.C.; Su, W.; Zhang, T. MicroRNA-10b Triggers the Epithelial-Mesenchymal Transition (EMT) of Laryngeal Carcinoma Hep-2 Cells by Directly Targeting the E-cadherin. Appl. Biochem. Biotechnol. 2015, 176, 33–44. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Uhlmann, E.J.; Wong, A.H.; Karmali, P.; Basu, M.; Gabriely, G.; Jain, A.; Wang, Y.; Chiocca, E.A.; Stephens, R.; et al. MicroRNA-10b inhibition reduces E2F1-mediated transcription and miR-15/16 activity in glioblastoma. Oncotarget 2015, 6, 3770–3783. [Google Scholar] [CrossRef] [PubMed]
- Biagioni, F.; Bossel Ben-Moshe, N.; Fontemaggi, G.; Canu, V.; Mori, F.; Antoniani, B.; di Benedetto, A.; Santoro, R.; Germoni, S.; de Angelis, F.; et al. miR-10b*, a master inhibitor of the cell cycle, is down-regulated in human breast tumours. EMBO Mol. Med. 2012, 4, 1214–1229. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.; Fu, Z.; Jiang, X.; Wang, N.; Wang, F.; Wang, X.; Zhang, S.; Wang, Y.; Yan, X.; Guan, W.X.; et al. miR-16 promotes the apoptosis of human cancer cells by targeting FEAT. BMC Cancer 2015, 15, 448. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Wang, Q.; Wang, G.D.; Wang, H.S.; Huang, Y.; Liu, X.M.; Cai, X.H. miR-16 inhibits cell proliferation by targeting IGF1R and the Raf1-MEK1/2-ERK1/2 pathway in osteosarcoma. FEBS Lett. 2013, 587, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.K.; Zhao, G.Y.; Tian, L.Y.; Liu, L.; Yan, K.; Ma, Y.L.; Ji, Z.W.; Li, X.X.; Han, K.; Gao, J.; et al. miR-15a and miR-16–1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncol. Rep. 2012, 28, 1764–1770. [Google Scholar] [PubMed]
- Yang, T.Q.; Lu, X.J.; Wu, T.F.; Ding, D.D.; Zhao, Z.H.; Chen, G.L.; Xie, X.S.; Li, B.; Wei, Y.X.; Guo, L.C.; et al. MicroRNA-16 inhibits glioma cell growth and invasion through suppression of BCL2 and the nuclear factor-κB1/MMP9 signaling pathway. Cancer Sci. 2014, 105, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xia, Y.; Niu, H.; Chen, Y. miR-16 induced the suppression of cell apoptosis while promote proliferation in esophageal squamous cell carcinoma. Cell. Physiol. Biochem. 2014, 33, 1340–1348. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Ren, X.; Chen, J.; Li, Y.; Tang, X.; Wen, X.; Yang, X.; Zhang, J.; Wang, Y.; Ma, J.; et al. miR-16 targets fibroblast growth factor 2 to inhibit NPC cell proliferation and invasion via PI3K/AKT and MAPK signaling pathways. Oncotarget 2015. [Google Scholar] [CrossRef]
- Zubillaga-Guerrero, M.I.; Alarcon-Romero Ldel, C.; Illades-Aguiar, B.; Flores-Alfaro, E.; Bermudez-Morales, V.H.; Deas, J.; Peralta-Zaragoza, O. MicroRNA miR-16-1 regulates CCNE1 (cyclin E1) gene expression in human cervical cancer cells. Int. J. Clin. Exp. Med. 2015, 8, 15999–16006. [Google Scholar] [PubMed]
- Ofir, M.; Hacohen, D.; Ginsberg, D. miR-15 and miR-16 are direct transcriptional targets of E2F1 that limit E2F-induced proliferation by targeting cyclin E. Mol. Cancer Res. 2011, 9, 440–447. [Google Scholar] [CrossRef] [PubMed]
- Huang, N.; Wu, J.; Qiu, W.; Lyu, Q.; He, J.; Xie, W.; Xu, N.; Zhang, Y. miR-15a and miR-16 induce autophagy and enhance chemosensitivity of Camptothecin. Cancer Biol. Ther. 2015, 16, 941–948. [Google Scholar] [CrossRef] [PubMed]
- Renjie, W.; Haiqian, L. miR-132, miR-15a and miR-16 synergistically inhibit pituitary tumor cell proliferation, invasion and migration by targeting Sox5. Cancer Lett. 2015, 356, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Shih, I.H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of mammalian microRNA targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef]
- Takakura, S.; Mitsutake, N.; Nakashima, M.; Namba, H.; Saenko, V.A.; Rogounovitch, T.I.; Nakazawa, Y.; Hayashi, T.; Ohtsuru, A.; Yamashita, S. Oncogenic role of miR-17-92 cluster in anaplastic thyroid cancer cells. Cancer Sci. 2008, 99, 1147–1154. [Google Scholar] [CrossRef] [PubMed]
- Dews, M.; Fox, J.L.; Hultine, S.; Sundaram, P.; Wang, W.; Liu, Y.Y.; Furth, E.; Enders, G.H.; El-Deiry, W.; Schelter, J.M.; et al. The myc-miR-17~92 axis blunts TGFβ signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010, 70, 8233–8246. [Google Scholar] [CrossRef] [PubMed]
- Mestdagh, P.; Bostrom, A.K.; Impens, F.; Fredlund, E.; van Peer, G.; de Antonellis, P.; von Stedingk, K.; Ghesquiere, B.; Schulte, S.; Dews, M.; et al. The miR-17–92 microRNA cluster regulates multiple components of the TGF-β pathway in neuroblastoma. Mol. Cell 2010, 40, 762–773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrocca, F.; Vecchione, A.; Croce, C.M. Emerging role of miR-106b-25/miR-17–92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 2008, 68, 8191–8194. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, V.; Fiorino, A.; Zoni, E.; Crippa, E.; Reid, J.F.; Gariboldi, M.; Pierotti, M.A. The Effects of miR-20a on p21: Two Mechanisms Blocking Growth Arrest in TGF-β-Responsive Colon Carcinoma. J. Cell. Physiol. 2015, 230, 3105–3114. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Lai, M.; Chen, M.; Xie, C.; Liao, R.; Kang, Y.J.; Xiao, C.; Hu, W.Y.; Han, J.; Sun, P. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010, 70, 8547–8557. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.J.; Li, J.S.; Zhou, H.; Xiao, H.X.; Li, Y.; Zhou, T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J. Exp. Clin. Cancer Res. 2015, 34, 73. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, Q.; Zhang, D.; Li, X.; Han, L.; Lei, J.; Duan, W.; Ma, Q.; Wu, Z.; Wang, Z. Upregulated miR-106a plays an oncogenic role in pancreatic cancer. FEBS Lett. 2014, 588, 705–712. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, B.; Shi, Y.; Xu, C.; Xiao, H.L.; Ma, L.N.; Xu, S.L.; Yang, L.; Wang, Q.L.; Dang, W.Q.; et al. Oncogenic miR-20a and miR-106a enhance the invasiveness of human glioma stem cells by directly targeting TIMP-2. Oncogene 2015, 34, 1407–1419. [Google Scholar] [CrossRef] [PubMed]
- Woods, K.; Thomson, J.M.; Hammond, S.M. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 2007, 282, 2130–2134. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.; Katiyar, S.K. Down-regulation of miRNA-106b inhibits growth of melanoma cells by promoting G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1 protein. Oncotarget 2014, 5, 10636–10649. [Google Scholar] [CrossRef] [PubMed]
- Dews, M.; Homayouni, A.; Yu, D.; Murphy, D.; Sevignani, C.; Wentzel, E.; Furth, E.E.; Lee, W.M.; Enders, G.H.; Mendell, J.T.; et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 2006, 38, 1060–1065. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Du, L.; Yang, X.; Zhang, X.; Wang, L.; Yang, Y.; Li, J.; Wang, C. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br. J. Cancer 2014, 111, 1985–1992. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, K.; Umezu, T.; Yoshizawa, S.; Ito, Y.; Ohyashiki, M.; Kawashima, H.; Tanaka, M.; Kuroda, M.; Ohyashiki, J.H. Clinical impact of down-regulated plasma miR-92a levels in non-Hodgkin’s lymphoma. PLoS ONE 2011, 6, e16408. [Google Scholar] [CrossRef] [PubMed]
- Yoshizawa, S.; Ohyashiki, J.H.; Ohyashiki, M.; Umezu, T.; Suzuki, K.; Inagaki, A.; Iida, S.; Ohyashiki, K. Downregulated plasma miR-92a levels have clinical impact on multiple myeloma and related disorders. Blood Cancer J. 2012, 2, e53. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Han, J.; Chen, J.; Dong, J.; Xia, Y.; Liu, J.; Jiang, Y.; Dai, J.; Lu, J.; Jin, G.; et al. Plasma miRNAs as early biomarkers for detecting hepatocellular carcinoma. Int. J. Cancer 2015, 137, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Buscaglia, L.E.; Li, Y. Apoptosis and the target genes of microRNA-21. Chin. J. Cancer 2011, 30, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Cmarik, J.L.; Min, H.; Hegamyer, G.; Zhan, S.; Kulesz-Martin, M.; Yoshinaga, H.; Matsuhashi, S.; Colburn, N.H. Differentially expressed protein Pdcd4 inhibits tumor promoter-induced neoplastic transformation. Proc. Natl. Acad. Sci. USA 1999, 96, 14037–14042. [Google Scholar] [CrossRef] [PubMed]
- Jansen, A.P.; Camalier, C.E.; Colburn, N.H. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005, 65, 6034–6041. [Google Scholar] [CrossRef] [PubMed]
- Leupold, J.H.; Yang, H.S.; Colburn, N.H.; Asangani, I.; Post, S.; Allgayer, H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene 2007, 26, 4550–4562. [Google Scholar] [CrossRef] [PubMed]
- Papagiannakopoulos, T.; Shapiro, A.; Kosik, K.S. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008, 68, 8164–8172. [Google Scholar] [CrossRef] [PubMed]
- Wickramasinghe, N.S.; Manavalan, T.T.; Dougherty, S.M.; Riggs, K.A.; Li, Y.; Klinge, C.M. Estradiol downregulates miR-21 expression and increases miR-21 target gene expression in MCF-7 breast cancer cells. Nucleic Acids Res. 2009, 37, 2584–2595. [Google Scholar] [PubMed]
- Meng, F.; Henson, R.; Wehbe-Janek, H.; Ghoshal, K.; Jacob, S.T.; Patel, T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology 2007, 133, 647–658. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Dong, X.; Zhai, B.; Jiang, X.; Dong, D.; Li, B.; Jiang, H.; Xu, S.; Sun, X. miR-21 mediates sorafenib resistance of hepatocellular carcinoma cells by inhibiting autophagy via the PTEN/Akt pathway. Oncotarget 2015, 6, 28867–28881. [Google Scholar] [PubMed]
- Gabriely, G.; Wurdinger, T.; Kesari, S.; Esau, C.C.; Burchard, J.; Linsley, P.S.; Krichevsky, A.M. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 2008, 28, 5369–5380. [Google Scholar] [CrossRef] [PubMed]
- Sabatel, C.; Malvaux, L.; Bovy, N.; Deroanne, C.; Lambert, V.; Gonzalez, M.L.; Colige, A.; Rakic, J.M.; Noel, A.; Martial, J.A.; et al. MicroRNA-21 exhibits antiangiogenic function by targeting RhoB expression in endothelial cells. PLoS ONE 2011, 6, e16979. [Google Scholar] [CrossRef] [PubMed]
- Zhu, S.; Si, M.L.; Wu, H.; Mo, Y.Y. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J. Biol. Chem. 2007, 282, 14328–14336. [Google Scholar] [CrossRef] [PubMed]
- Garzon, R.; Heaphy, C.E.; Havelange, V.; Fabbri, M.; Volinia, S.; Tsao, T.; Zanesi, N.; Kornblau, S.M.; Marcucci, G.; Calin, G.A.; et al. MicroRNA 29b functions in acute myeloid leukemia. Blood 2009, 114, 5331–5341. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, L.H.; Schiavinato, J.L.; Fraguas, M.S.; Lucena-Araujo, A.R.; Haddad, R.; Araujo, A.G.; Dalmazzo, L.F.; Rego, E.M.; Covas, D.T.; Zago, M.A.; et al. Potential roles of microRNA-29a in the molecular pathophysiology of T-cell acute lymphoblastic leukemia. Cancer Sci. 2015, 106, 1264–1277. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Xu, Y.; Jing, Z.; Wang, X.; Zha, X.; Zeng, C.; Chen, S.; Yang, L.; Luo, G.; Li, B.; et al. Altered expression pattern of miR-29a, miR-29b and the target genes in myeloid leukemia. Exp. Hematol. Oncol. 2014, 3, 17. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Kuo, C.; Nicholl, M.B.; Sim, M.S.; Turner, R.R.; Morton, D.L.; Hoon, D.S. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics 2011, 6, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.L.; Wang, W.; Hu, Z.; Wang, L.W.; Chang, J.; Qian, H. Negative feedback of miR-29 family TET1 involves in hepatocellular cancer. Med. Oncol. 2014, 31, 291. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Fang, J.H.; Yun, J.P.; Yang, J.; Zhang, Y.; Jia, W.H.; Zhuang, S.M. Effects of microRNA-29 on apoptosis, tumorigenicity, and prognosis of hepatocellular carcinoma. Hepatology 2010, 51, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Inoue, A.; Yamamoto, H.; Uemura, M.; Nishimura, J.; Hata, T.; Takemasa, I.; Ikenaga, M.; Ikeda, M.; Murata, K.; Mizushima, T.; et al. MicroRNA-29b is a Novel Prognostic Marker in Colorectal Cancer. Ann. Surg. Oncol. 2015, 22, 1410–1418. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, F.; Xu, J.; Ye, F.; Shen, Y.; Zhou, J.; Lu, W.; Wan, X.; Ma, D.; Xie, X. Progressive miRNA expression profiles in cervical carcinogenesis and identification of HPV-related target genes for miR-29. J. Pathol. 2011, 224, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.W.; Hsu, N.Y.; Wang, Y.C.; Lee, M.C.; Cheng, Y.W.; Chen, C.Y.; Lee, H. c-Myc suppresses microRNA-29b to promote tumor aggressiveness and poor outcomes in non-small cell lung cancer by targeting FHIT. Oncogene 2015, 34, 2072–2082. [Google Scholar] [CrossRef] [PubMed]
- Nishikawa, R.; Goto, Y.; Kojima, S.; Enokida, H.; Chiyomaru, T.; Kinoshita, T.; Sakamoto, S.; Fuse, M.; Nakagawa, M.; Naya, Y.; et al. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int. J. Oncol. 2014, 45, 401–410. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.J.; Lin, J.; Lwin, T.; Yang, H.; Guo, J.; Kong, W.; Dessureault, S.; Moscinski, L.C.; Rezania, D.; Dalton, W.S.; et al. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood 2010, 115, 2630–2639. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, J.H.; Ha, M.; Nam, J.W.; Kim, V.N. MiR-29 miRNAs activate p53 by targeting p85 α and CDC42. Nat. Struct. Mol. Biol. 2009, 16, 23–29. [Google Scholar] [CrossRef] [PubMed]
- Ye, Z.; Zhao, L.; Li, J.; Chen, W.; Li, X. MiR-30d Blocked Transforming Growth Factor β1-Induced Epithelial-Mesenchymal Transition by Targeting Snail in Ovarian Cancer Cells. Int. J. Gynecol. Cancer 2015, 25, 1574–1581. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Wang, X.; Huo, Q.; Sun, M.; Cai, C.; Liu, Z.; Hu, G.; Yang, Q. MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 2014, 33, 3119–3128. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.Y.; Zhang, X.C.; Su, J.; Meng, W.; Yang, X.N.; Yang, J.J.; Zhou, Q.; Chen, Z.Y.; Chen, Z.H.; Xie, Z.; et al. BCL11A overexpression predicts survival and relapse in non-small cell lung cancer and is modulated by microRNA-30a and gene amplification. Mol. Cancer 2013, 12, 61. [Google Scholar] [CrossRef] [PubMed]
- Zhong, M.; Bian, Z.; Wu, Z. miR-30a suppresses cell migration and invasion through downregulation of PIK3CD in colorectal carcinoma. Cell. Physiol. Biochem. 2013, 31, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Zhong, K.; Chen, K.; Han, L.; Li, B. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer 2014, 14, 703. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, Q.; Qin, D.; Yu, L.; Huang, R.; Lv, G.; Zou, Z.; Jiang, X.C.; Zou, C.; Liu, W.; et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol. Cell. Biol. 2015, 35, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, F.; Tang, S. MiR-30a upregulates BCL2A1, IER3 and cyclin D2 expression by targeting FOXL2. Oncol. Lett. 2015, 9, 967–971. [Google Scholar] [PubMed]
- Kao, C.J.; Martiniez, A.; Shi, X.B.; Yang, J.; Evans, C.P.; Dobi, A.; deVere White, R.W.; Kung, H.J. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene 2014, 33, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Ouzounova, M.; Vuong, T.; Ancey, P.B.; Ferrand, M.; Durand, G.; Le-Calvez Kelm, F.; Croce, C.; Matar, C.; Herceg, Z.; Hernandez-Vargas, H. MicroRNA miR-30 family regulates non-attachment growth of breast cancer cells. BMC Genom. 2013, 14, 139. [Google Scholar] [CrossRef] [PubMed]
- Bridge, G.; Monteiro, R.; Henderson, S.; Emuss, V.; Lagos, D.; Georgopoulou, D.; Patient, R.; Boshoff, C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood 2012, 120, 5063–5072. [Google Scholar] [CrossRef] [PubMed]
- Maroof, H.; Salajegheh, A.; Smith, R.A.; Lam, A.K. Role of microRNA-34 family in cancer with particular reference to cancer angiogenesis. Exp. Mol. Pathol. 2014, 97, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Ghandadi, M.; Sahebkar, A. MicroRNA-34a and its target genes: Key factors in cancer multidrug resistance. Curr. Pharm. Des. 2016, 22, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.; Lieberman, J. miR-34 and p53: New Insights into a Complex Functional Relationship. PLoS ONE 2015, 10, e0132767. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.J.; Lin, C.P.; Ho, J.J.; He, X.; Okada, N.; Bu, P.; Zhong, Y.; Kim, S.Y.; Bennett, M.J.; Chen, C.; et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat. Cell Biol. 2011, 13, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- De Antonellis, P.; Medaglia, C.; Cusanelli, E.; Andolfo, I.; Liguori, L.; de Vita, G.; Carotenuto, M.; Bello, A.; Formiggini, F.; Galeone, A.; et al. miR-34a targeting of Notch ligand delta-like 1 impairs CD15+/CD133+ tumor-propagating cells and supports neural differentiation in medulloblastoma. PLoS ONE 2011, 6, e24584. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.H.; Kim, N.H.; Park, C.; Lee, I.; Kim, H.S.; Yook, J.I. MiRNA-34 intrinsically links p53 tumor suppressor and Wnt signaling. Cell Cycle 2012, 11, 1273–1281. [Google Scholar] [CrossRef] [PubMed]
- Siemens, H.; Jackstadt, R.; Hunten, S.; Kaller, M.; Menssen, A.; Gotz, U.; Hermeking, H. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 2011, 10, 4256–4271. [Google Scholar] [CrossRef] [PubMed]
- Brzozowa, M.; Mielanczyk, L.; Michalski, M.; Malinowski, L.; Kowalczyk-Ziomek, G.; Helewski, K.; Harabin-Slowinska, M.; Wojnicz, R. Role of Notch signaling pathway in gastric cancer pathogenesis. Contemp. Oncol. (Pozn) 2013, 17, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Bae, Y.; Yang, T.; Zeng, H.C.; Campeau, P.M.; Chen, Y.; Bertin, T.; Dawson, B.C.; Munivez, E.; Tao, J.; Lee, B.H. miRNA-34c regulates Notch signaling during bone development. Hum. Mol. Genet. 2012, 21, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Yao, H.; Zheng, Z.; Qiu, G.; Sun, K. miR-125b targets BCL3 and suppresses ovarian cancer proliferation. Int. J. Cancer 2011, 128, 2274–2283. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Luo, J.; Cai, Q.; Pan, Q.; Zeng, H.; Guo, Z.; Dong, W.; Huang, J.; Lin, T. MicroRNA-125b suppresses the development of bladder cancer by targeting E2F3. Int. J. Cancer 2011, 128, 1758–1769. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Duan, R.; Kooy, F.; Sherman, S.L.; Zhou, W.; Jin, P. Germline mutation of microRNA-125a is associated with breast cancer. J. Med. Genet. 2009, 46, 358–360. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Liu, Z.; Zhao, Y.; Ding, Y.; Liu, H.; Xi, Y.; Xiong, W.; Li, G.; Lu, J.; Fodstad, O.; et al. MicroRNA-125b confers the resistance of breast cancer cells to paclitaxel through suppression of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J. Biol. Chem. 2010, 285, 21496–21507. [Google Scholar] [CrossRef] [PubMed]
- Bi, Q.; Tang, S.; Xia, L.; Du, R.; Fan, R.; Gao, L.; Jin, J.; Liang, S.; Chen, Z.; Xu, G.; et al. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS ONE 2012, 7, e40169. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Wong, C.M.; Ying, Q.; Fan, D.N.; Huang, S.; Ding, J.; Yao, J.; Yan, M.; Li, J.; Yao, M.; et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology 2010, 52, 1731–1740. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.Y.; Wang, Y.X.; Yan, W.T.; Li, H.Y.; Tian, Y.Z.; Wang, S.M.; Zhao, H.L. MicroRNA-125b functions as a tumor suppressor in hepatocellular carcinoma cells. Int. J. Mol. Sci. 2012, 13, 8762–8774. [Google Scholar] [CrossRef] [PubMed]
- Kappelmann, M.; Kuphal, S.; Meister, G.; Vardimon, L.; Bosserhoff, A.K. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2013, 32, 2984–2991. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Zhang, L.; Meisgen, F.; Harada, M.; Heilborn, J.; Homey, B.; Grander, D.; Stahle, M.; Sonkoly, E.; Pivarcsi, A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J. Biol. Chem. 2012, 287, 29899–29908. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.H.; Li, H.; Li, J.P.; Zhong, H.; Zhang, H.C.; Chen, J.; Xiao, T. miR-125b suppresses the proliferation and migration of osteosarcoma cells through down-regulation of STAT3. Biochem. Biophys. Res. Commun. 2011, 416, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Zhang, J.P.; Li, B.; Zeng, C.; You, K.; Chen, M.X.; Yuan, Y.; Zhuang, S.M. MicroRNA-125b promotes apoptosis by regulating the expression of Mcl-1, Bcl-w and IL-6R. Oncogene 2013, 32, 3071–3079. [Google Scholar] [CrossRef] [PubMed]
- Zhao, A.; Zeng, Q.; Xie, X.; Zhou, J.; Yue, W.; Li, Y.; Pei, X. MicroRNA-125b induces cancer cell apoptosis through suppression of Bcl-2 expression. J. Genet. Genom. 2012, 39, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Young, C.Y.; Yuan, H. MicroRNAs and prostate cancer. Acta Biochim. Biophys. Sin. (Shanghai) 2010, 42, 363–369. [Google Scholar] [CrossRef] [PubMed]
- Mashima, R. Physiological roles of miR-155. Immunology 2015, 145, 323–333. [Google Scholar] [CrossRef] [PubMed]
- Seddiki, N.; Brezar, V.; Ruffin, N.; Levy, Y.; Swaminathan, S. Role of miR-155 in the regulation of lymphocyte immune function and disease. Immunology 2014, 142, 32–38. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Lewis, M.G.; Renne, R.; Mattapallil, J.J. Suppression of transforming growth factor beta receptor 2 and Smad5 is associated with high levels of microRNA miR-155 in the oral mucosa during chronic simian immunodeficiency virus infection. J. Virol. 2015, 89, 2972–2978. [Google Scholar] [CrossRef] [PubMed]
- Yao, R.; Ma, Y.L.; Liang, W.; Li, H.H.; Ma, Z.J.; Yu, X.; Liao, Y.H. MicroRNA-155 modulates Treg and Th17 cells differentiation and Th17 cell function by targeting SOCS1. PLoS ONE 2012, 7, e46082. [Google Scholar] [CrossRef] [PubMed]
- Xue, H.; Hua, L.M.; Guo, M.; Luo, J.M. SHIP1 is targeted by miR-155 in acute myeloid leukemia. Oncol. Rep. 2014, 32, 2253–2259. [Google Scholar] [CrossRef] [PubMed]
- Salemi, D.; Cammarata, G.; Agueli, C.; Augugliaro, L.; Corrado, C.; Bica, M.G.; Raimondo, S.; Marfia, A.; Randazzo, V.; Dragotto, P.; et al. miR-155 regulative network in FLT3 mutated acute myeloid leukemia. Leuk. Res. 2015, 39, 883–896. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Zhu, M.; Corbalan-Campos, J.; Heyll, K.; Weber, C.; Schober, A. Regulation of Csf1r and Bcl6 in macrophages mediates the stage-specific effects of microRNA-155 on atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 796–803. [Google Scholar] [CrossRef] [PubMed]
- De Santis, R.; Liepelt, A.; Mossanen, J.C.; Dueck, A.; Simons, N.; Mohs, A.; Trautwein, C.; Meister, G.; Marx, G.; Ostareck-Lederer, A.; et al. miR-155 targets Caspase-3 mRNA in activated macrophages. RNA Biol. 2016, 13, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Q.; Yu, X.D.; Liu, Z.H.; Cheng, X.; Samartzis, D.; Jia, L.T.; Wu, S.X.; Huang, J.; Chen, J.; Luo, Z.J. Deregulated miR-155 promotes Fas-mediated apoptosis in human intervertebral disc degeneration by targeting FADD and caspase-3. J. Pathol. 2011, 225, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.S.; Zhong, Y.F.; Fang, Z.; Li, B.; An, J. miR-155 inhibits the sensitivity of lung cancer cells to cisplatin via negative regulation of Apaf-1 expression. Cancer Gene Ther. 2012, 19, 773–778. [Google Scholar] [CrossRef] [PubMed]
- Ling, N.; Gu, J.; Lei, Z.; Li, M.; Zhao, J.; Zhang, H.T.; Li, X. MicroRNA-155 regulates cell proliferation and invasion by targeting FOXO3a in glioma. Oncol. Rep. 2013, 30, 2111–2118. [Google Scholar] [PubMed]
- Azimzadeh, M.; Rahaie, M.; Nasirizadeh, N.; Ashtari, K.; Naderi-Manesh, H. An electrochemical nanobiosensor for plasma miRNA-155, based on graphene oxide and gold nanorod, for early detection of breast cancer. Biosens. Bioelectron. 2016, 77, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Paik, W.H.; Song, B.J.; Kim, H.W.; Kim, H.R.; Hwang, J.H. MicroRNA-200c as a Prognostic Biomarker for Pancreatic Cancer. Korean J. Gastroenterol. 2015, 66, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Zaravinos, A. The Regulatory Role of MicroRNAs in EMT and Cancer. J. Oncol. 2015, 2015, 865816. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.C.; Yoon, S.; Jeong, Y.; Yoon, J.; Baek, K. Regulation of vascular endothelial growth factor signaling by miR-200b. Mol. Cells 2011, 32, 77–82. [Google Scholar] [CrossRef] [PubMed]
- McArthur, K.; Feng, B.; Wu, Y.; Chen, S.; Chakrabarti, S. MicroRNA-200b regulates vascular endothelial growth factor-mediated alterations in diabetic retinopathy. Diabetes 2011, 60, 1314–1323. [Google Scholar] [CrossRef] [PubMed]
- Pecot, C.V.; Rupaimoole, R.; Yang, D.; Akbani, R.; Ivan, C.; Lu, C.; Wu, S.; Han, H.D.; Shah, M.Y.; Rodriguez-Aguayo, C.; et al. Tumour angiogenesis regulation by the miR-200 family. Nat. Commun. 2013, 4, 2427. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Xie, H.Y.; Zhu, C.D.; Zhu, X.F.; Cao, G.X.; Chen, X.H.; Xu, H.F. Increased miR-141 expression is associated with diagnosis and favorable prognosis of patients with bladder cancer. Tumour Biol. 2015, 36, 877–883. [Google Scholar] [CrossRef] [PubMed]
- Fasanaro, P.; D’Alessandra, Y.; di Stefano, V.; Melchionna, R.; Romani, S.; Pompilio, G.; Capogrossi, M.C.; Martelli, F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J. Biol. Chem. 2008, 283, 15878–15883. [Google Scholar] [CrossRef] [PubMed]
- Pulkkinen, K.; Malm, T.; Turunen, M.; Koistinaho, J.; Yla-Herttuala, S. Hypoxia induces microRNA miR-210 in vitro and in vivo ephrin-A3 and neuronal pentraxin 1 are potentially regulated by miR-210. FEBS Lett. 2008, 582, 2397–2401. [Google Scholar] [CrossRef] [PubMed]
- Foekens, J.A.; Sieuwerts, A.M.; Smid, M.; Look, M.P.; de Weerd, V.; Boersma, A.W.; Klijn, J.G.; Wiemer, E.A.; Martens, J.W. Four miRNAs associated with aggressiveness of lymph node-negative, estrogen receptor-positive human breast cancer. Proc. Natl. Acad. Sci. USA 2008, 105, 13021–13026. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, M.; Nasu, K.; Abe, W.; Aoyagi, Y.; Kawano, Y.; Kai, K.; Moriyama, M.; Narahara, H. Enhanced miR-210 expression promotes the pathogenesis of endometriosis through activation of signal transducer and activator of transcription 3. Hum. Reprod. 2015, 30, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sun, H.; Dai, H.; Walsh, R.M.; Imakura, M.; Schelter, J.; Burchard, J.; Dai, X.; Chang, A.N.; Diaz, R.L.; et al. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle 2009, 8, 2756–2768. [Google Scholar] [CrossRef] [PubMed]
- Giannakakis, A.; Sandaltzopoulos, R.; Greshock, J.; Liang, S.; Huang, J.; Hasegawa, K.; Li, C.; O’Brien-Jenkins, A.; Katsaros, D.; Weber, B.L.; et al. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol. Ther. 2008, 7, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Nakada, C.; Tsukamoto, Y.; Matsuura, K.; Nguyen, T.L.; Hijiya, N.; Uchida, T.; Sato, F.; Mimata, H.; Seto, M.; Moriyama, M. Overexpression of miR-210, a downstream target of HIF1α, causes centrosome amplification in renal carcinoma cells. J. Pathol. 2011, 224, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Tsuchiya, S.; Fujiwara, T.; Sato, F.; Shimada, Y.; Tanaka, E.; Sakai, Y.; Shimizu, K.; Tsujimoto, G. MicroRNA-210 regulates cancer cell proliferation through targeting fibroblast growth factor receptor-like 1 (FGFRL1). J. Biol. Chem. 2011, 286, 420–428. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhou, X.Y.; Zhou, X.G.; Cheng, R.; Liu, H.Y.; Li, Y. Neuroprotective effects of microRNA-210 against oxygen-glucose deprivation through inhibition of apoptosis in PC12 cells. Mol. Med. Rep. 2013, 7, 1955–1959. [Google Scholar] [CrossRef] [PubMed]
- Chio, C.C.; Lin, J.W.; Cheng, H.A.; Chiu, W.T.; Wang, Y.H.; Wang, J.J.; Hsing, C.H.; Chen, R.M. MicroRNA-210 targets antiapoptotic Bcl-2 expression and mediates hypoxia-induced apoptosis of neuroblastoma cells. Arch. Toxicol. 2013, 87, 459–468. [Google Scholar] [CrossRef] [PubMed]
- Crosby, M.E.; Kulshreshtha, R.; Ivan, M.; Glazer, P.M. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009, 69, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Ding, L.; Bennewith, K.L.; Tong, R.T.; Welford, S.M.; Ang, K.K.; Story, M.; Le, Q.T.; Giaccia, A.J. Hypoxia-inducible miR-210 regulates normoxic gene expression involved in tumor initiation. Mol. Cell 2009, 35, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Urbich, C.; Kuehbacher, A.; Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 2008, 79, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Poliseno, L.; Tuccoli, A.; Mariani, L.; Evangelista, M.; Citti, L.; Woods, K.; Mercatanti, A.; Hammond, S.; Rainaldi, G. MicroRNAs modulate the angiogenic properties of HUVECs. Blood 2006, 108, 3068–3071. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.; Bai, W.; Zhu, H.; Zhang, X.; Chen, Y.; Wang, L.; Yang, A.; Zhao, J.; Jia, L. miR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014, 47, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Di Leva, G.; Romano, G.; Nuovo, G.; Suh, S.S.; Ngankeu, A.; Taccioli, C.; Pichiorri, F.; Alder, H.; Secchiero, P.; et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16, 498–509. [Google Scholar] [PubMed]
- Suarez, Y.; Fernandez-Hernando, C.; Pober, J.S.; Sessa, W.C. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007, 100, 1164–1173. [Google Scholar] [CrossRef] [PubMed]
- Cai, G.; Qiao, S.; Chen, K. Suppression of miR-221 inhibits glioma cells proliferation and invasion via targeting SEMA3B. Biol. Res. 2015, 48, 37. [Google Scholar] [CrossRef] [PubMed]
- Kneitz, B.; Krebs, M.; Kalogirou, C.; Schubert, M.; Joniau, S.; van Poppel, H.; Lerut, E.; Kneitz, S.; Scholz, C.J.; Strobel, P.; et al. Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res. 2014, 74, 2591–2603. [Google Scholar] [CrossRef] [PubMed]
- Zhong, C.; Ding, S.; Xu, Y.; Huang, H. MicroRNA-222 promotes human non-small cell lung cancer H460 growth by targeting p27. Int. J. Clin. Exp. Med. 2015, 8, 5534–5540. [Google Scholar] [PubMed]
- Yang, C.J.; Shen, W.G.; Liu, C.J.; Chen, Y.W.; Lu, H.H.; Tsai, M.M.; Lin, S.C. MiR-221 and miR-222 expression increased the growth and tumorigenesis of oral carcinoma cells. J. Oral Pathol. Med. 2011, 40, 560–566. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wang, X.; He, H.H.; Sweeney, C.J.; Liu, S.X.; Brown, M.; Balk, S.; Lee, G.S.; Kantoff, P.W. miR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene 2014, 33, 2790–2800. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.; Di Leva, G.; Li, M.; Fang, F.; Devlin, C.; Hartman-Frey, C.; Burow, M.E.; Ivan, M.; Croce, C.M.; Nephew, K.P. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011, 30, 1082–1097. [Google Scholar] [CrossRef] [PubMed]
- Dentelli, P.; Traversa, M.; Rosso, A.; Togliatto, G.; Olgasi, C.; Marchio, C.; Provero, P.; Lembo, A.; Bon, G.; Annaratone, L.; et al. miR-221/222 control luminal breast cancer tumor progression by regulating different targets. Cell Cycle 2014, 13, 1811–1826. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Zhang, J.X.; Zhang, A.L.; Shi, Z.D.; Han, L.; Jia, Z.F.; Yang, W.D.; Wang, G.X.; Jiang, T.; You, Y.P.; et al. miR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol. Cancer 2010, 9, 229. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Kurozumi, A.; Kato, M.; Enokida, H.; Matsushita, R.; Yamazaki, K.; Ishida, Y.; Nakagawa, M.; et al. MicroRNA expression signature of castration-resistant prostate cancer: The microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 2015, 113, 1055–1065. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Xu, Y.; Zhang, W.; Deng, Y.; Si, M.; Du, Y.; Yao, H.; Liu, X.; Ke, Y.; Si, J.; et al. miR-375 frequently downregulated in gastric cancer inhibits cell proliferation by targeting JAK2. Cell Res. 2010, 20, 784–793. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, Y.; Nakada, C.; Noguchi, T.; Tanigawa, M.; Nguyen, L.T.; Uchida, T.; Hijiya, N.; Matsuura, K.; Fujioka, T.; Seto, M.; et al. MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res. 2010, 70, 2339–2349. [Google Scholar] [CrossRef] [PubMed]
- Song, S.D.; Zhou, J.; Zhou, J.; Zhao, H.; Cen, J.N.; Li, D.C. MicroRNA-375 targets the 3-phosphoinositide-dependent protein kinase-1 gene in pancreatic carcinoma. Oncol. Lett. 2013, 6, 953–959. [Google Scholar] [PubMed]
- Luo, J.; Wu, J.; Li, Z.; Qin, H.; Wang, B.; Wong, T.S.; Yang, W.; Fu, Q.L.; Lei, W. miR-375 suppresses IGF1R expression and contributes to inhibition of cell progression in laryngeal squamous cell carcinoma. BioMed Res. Int. 2014, 2014, 374598. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Quan, T.; Luo, B.; Guo, X.; Liu, L.; Zheng, Q. MiR-375 targets KLF4 and impacts the proliferation of colorectal carcinoma. Tumour Biol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Yang, J.; Li, S.; Shan, X.; Liu, X.; Hua, H.; Zhao, C.; Feng, Z.; Cai, Z.; Zhang, L.; et al. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget 2015, 6, 40172–40185. [Google Scholar] [PubMed]
- Wang, X.Z.; Hang, Y.K.; Liu, J.B.; Hou, Y.Q.; Wang, N.; Wang, M.J. Over-expression of microRNA-375 inhibits papillary thyroid carcinoma cell proliferation and induces cell apoptosis by targeting ERBB2. J. Pharmacol. Sci. 2016, 130, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.Y.; Zhang, Z.Z.; Liu, H.; Zhao, E.H.; Cao, H. miR-375 inhibits the proliferation of gastric cancer cells by repressing ERBB2 expression. Exp. Ther. Med. 2014, 7, 1757–1761. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, Q.; Li, M.; Jiang, S.; Wang, X. MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem. Biophys. Res. Commun. 2014, 444, 199–204. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cao, Y.; Su, T.; Jiang, Y.; Jiang, L.; Zhou, W.; Zhang, C.; Wang, W.; Ning, G. Downregulation of miR-375 in aldosterone-producing adenomas promotes tumour cell growth via MTDH. Clin. Endocrinol. (Oxf.) 2015, 83, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Hudson, J.; Duncavage, E.; Tamburrino, A.; Salerno, P.; Xi, L.; Raffeld, M.; Moley, J.; Chernock, R.D. Overexpression of miR-10a and miR-375 and downregulation of YAP1 in medullary thyroid carcinoma. Exp. Mol. Pathol. 2013, 95, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.M.; Patel, R.S.; Phillips, B.L.; Wang, H.; Cohen, D.M.; Reinhold, W.C.; Chang, L.J.; Yang, L.J.; Chan, E.K. Tumor suppressor miR-375 regulates MYC expression via repression of CIP2A coding sequence through multiple miRNA-mRNA interactions. Mol. Biol. Cell 2013, 24, 1638–1648. [Google Scholar] [CrossRef] [PubMed]
- He, X.X.; Chang, Y.; Meng, F.Y.; Wang, M.Y.; Xie, Q.H.; Tang, F.; Li, P.Y.; Song, Y.H.; Lin, J.S. MicroRNA-375 targets AEG-1 in hepatocellular carcinoma and suppresses liver cancer cell growth in vitro and in vivo. Oncogene 2012, 31, 3357–3369. [Google Scholar] [CrossRef] [PubMed]
- Zaharie, F.; Muresan, M.S.; Petrushev, B.; Berce, C.; Gafencu, G.A.; Selicean, S.; Jurj, A.; Cojocneanu-Petric, R.; Lisencu, C.I.; Pop, L.A.; et al. Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. J. Gastroint. Liver Dis. 2015, 24, 435–443. [Google Scholar]
- Yoda, S.; Soejima, K.; Hamamoto, J.; Yasuda, H.; Nakayama, S.; Satomi, R.; Terai, H.; Ikemura, S.; Sato, T.; Naoki, K.; et al. Claudin-1 is a novel target of miR-375 in non-small-cell lung cancer. Lung Cancer 2014, 85, 366–372. [Google Scholar] [CrossRef] [PubMed]
- Ye, X.M.; Zhu, H.Y.; Bai, W.D.; Wang, T.; Wang, L.; Chen, Y.; Yang, A.G.; Jia, L.T. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer 2014, 14, 134. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xing, R.; Zhang, X.; Dong, W.; Zhang, J.; Yan, Z.; Li, W.; Cui, J.; Lu, Y. miR-375 targets the p53 gene to regulate cellular response to ionizing radiation and etoposide in gastric cancer cells. DNA Repair. (Amst.) 2013, 12, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Mussnich, P.; Rosa, R.; Bianco, R.; Fusco, A.; D'Angelo, D. miR-199a-5p and miR-375 affect colon cancer cell sensitivity to cetuximab by targeting PHLPP1. Expert Opin. Ther. Targets 2015, 19, 1017–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.; Noh, H.; Teng, Y.; Shao, J.; Rehmani, H.; Ding, H.F.; Dong, Z.; Su, S.B.; Shi, H.; Kim, J.; et al. SHOX2 is a direct miR-375 target and a novel epithelial-to-mesenchymal transition inducer in breast cancer cells. Neoplasia 2014, 16, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yan, W.; He, X.; Zhang, L.; Li, C.; Huang, H.; Nace, G.; Geller, D.A.; Lin, J.; Tsung, A. miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterology 2012, 143, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Szigeti, A.; Tatrai, E.; Szamosi, A.; Vargha, P.; Nagy, Z.Z.; Nemeth, J.; DeBuc, D.C.; Somfai, G.M. A morphological study of retinal changes in unilateral amblyopia using optical coherence tomography image segmentation. PLoS ONE 2014, 9, e88363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, N.J.; Zhou, X.; Yu, T.; Brinkman, B.M.; Zimmermann, B.G.; Palanisamy, V.; Wong, D.T. Characterization of salivary RNA by cDNA library analysis. Arch. Oral Biol. 2007, 52, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Lin, S.C.; Yang, C.C.; Cheng, H.W.; Chang, K.W. Exploiting salivary miR-31 as a clinical biomarker of oral squamous cell carcinoma. Head Neck 2012, 34, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Trachtenberg, A.J.; Kuo, W.P.; Cheng, Y.S. Genomewide Study of Salivary MicroRNAs for Detection of Oral Cancer. J. Dent. Res. 2014, 93, 86S–93S. [Google Scholar] [CrossRef] [PubMed]
- Zahran, F.; Ghalwash, D.; Shaker, O.; Al-Johani, K.; Scully, C. Salivary microRNAs in oral cancer. Oral Dis. 2015, 21, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Duz, M.B.; Karatas, O.F.; Guzel, E.; Turgut, N.F.; Yilmaz, M.; Creighton, C.J.; Ozen, M. Identification of miR-139-5p as a saliva biomarker for tongue squamous cell carcinoma: A pilot study. Cell. Oncol. (Dordr.) 2015. [Google Scholar] [CrossRef] [PubMed]
- Matse, J.H.; Yoshizawa, J.; Wang, X.; Elashoff, D.; Bolscher, J.G.; Veerman, E.C.; Bloemena, E.; Wong, D.T. Discovery and prevalidation of salivary extracellular microRNA biomarkers panel for the noninvasive detection of benign and malignant parotid gland tumors. Clin. Cancer Res. 2013, 19, 3032–3038. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Yin, X.; Gong, B.; Nie, W.; Wu, B.; Zhang, X.; Huang, J.; Zhang, P.; Zhou, Z.; Li, Z. Salivary microRNAs show potential as a noninvasive biomarker for detecting resectable pancreatic cancer. Cancer Prev. Res. (Phila) 2015, 8, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Liang, M.; Dittmar, R.; Wang, L. Extracellular microRNAs in urologic malignancies: Chances and challenges. Int. J. Mol. Sci. 2013, 14, 14785–14799. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Gong, G.; Tan, H.; Dai, F.; Zhu, X.; Chen, Y.; Wang, J.; Liu, Y.; Chen, P.; Wu, X.; et al. Urinary microRNA-30a-5p is a potential biomarker for ovarian serous adenocarcinoma. Oncol. Rep. 2015, 33, 2915–2923. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.A.; Haj-Ahmad, Y. Promising Candidate Urinary MicroRNA Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J. Cancer 2012, 3, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Machida, A.; Ohkubo, T.; Yokota, T. Circulating microRNAs in the cerebrospinal fluid of patients with brain diseases. Methods Mol. Biol. 2013, 1024, 203–209. [Google Scholar] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Chan, A.; Deckert, M.; Gold, R.; Maghnouj, A.; Zollner, H.; Reinacher-Schick, A.; Schmiegel, W.; et al. Identification of microRNAs in the cerebrospinal fluid as marker for primary diffuse large B-cell lymphoma of the central nervous system. Blood 2011, 117, 3140–3146. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Maghnouj, A.; Zollner, H.; Schmiegel, W.; Hahn, S.; Schroers, R. Identification of microRNAs in the cerebrospinal fluid as biomarker for the diagnosis of glioma. Neuro Oncol. 2012, 14, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Baraniskin, A.; Kuhnhenn, J.; Schlegel, U.; Schmiegel, W.; Hahn, S.; Schroers, R. MicroRNAs in cerebrospinal fluid as biomarker for disease course monitoring in primary central nervous system lymphoma. J. Neurooncol. 2012, 109, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Tuo, J.; Shen, D.; Yang, H.H.; Chan, C.C. Distinct microRNA-155 expression in the vitreous of patients with primary vitreoretinal lymphoma and uveitis. Am. J. Ophthalmol. 2014, 157, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Ragusa, M.; Caltabiano, R.; Russo, A.; Puzzo, L.; Avitabile, T.; Longo, A.; Toro, M.D.; di Pietro, C.; Purrello, M.; Reibaldi, M. MicroRNAs in vitreus humor from patients with ocular diseases. Mol. Vis. 2013, 19, 430–440. [Google Scholar] [PubMed]
- Gu, Y.Q.; Gong, G.; Xu, Z.L.; Wang, L.Y.; Fang, M.L.; Zhou, H.; Xing, H.; Wang, K.R.; Sun, L. miRNA profiling reveals a potential role of milk stasis in breast carcinogenesis. Int. J. Mol. Med. 2014, 33, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Roberts, M.J.; Chow, C.W.; Marshall, V.R.; Doi, S.A.; Vincent, A.D.; Butler, L.M.; Lavin, M.F.; Tilley, W.D.; Gardiner, R.A. Human seminal fluid as a source of prostate cancer-specific microRNA biomarkers. Endocr. Relat. Cancer 2014, 21, L17–L21. [Google Scholar] [CrossRef] [PubMed]
- Margue, C.; Reinsbach, S.; Philippidou, D.; Beaume, N.; Walters, C.; Schneider, J.G.; Nashan, D.; Behrmann, I.; Kreis, S. Comparison of a healthy miRNome with melanoma patient miRNomes: Are microRNAs suitable serum biomarkers for cancer? Oncotarget 2015, 6, 12110–12127. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef] [PubMed]
- McDonald, J.S.; Milosevic, D.; Reddi, H.V.; Grebe, S.K.; Algeciras-Schimnich, A. Analysis of circulating microRNA: Preanalytical and analytical challenges. Clin. Chem. 2011, 57, 833–840. [Google Scholar] [CrossRef] [PubMed]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during sample preparation alters microRNA content of plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, C.C.; Kroh, E.; Wood, B.; Arroyo, J.D.; Dougherty, K.J.; Miyaji, M.M.; Tait, J.F.; Tewari, M. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila) 2012, 5, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Kannan, M.; Atreya, C. Differential profiling of human red blood cells during storage for 52 selected microRNAs. Transfusion 2010, 50, 1581–1588. [Google Scholar] [CrossRef] [PubMed]
- Al-Soud, W.A.; Radstrom, P. Purification and characterization of PCR-inhibitory components in blood cells. J. Clin. Microbiol. 2001, 39, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; Kerin, M.J. Circulating miRNA signatures: Promising prognostic tools for cancer. J. Clin. Oncol. 2010, 28, e573–e574. [Google Scholar] [CrossRef] [PubMed]
- Gourzones, C.; Ferrand, F.R.; Amiel, C.; Verillaud, B.; Barat, A.; Guerin, M.; Gattolliat, C.H.; Gelin, A.; Klibi, J.; Chaaben, A.B.; et al. Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients—Evidence of non-exosomal transport. Virol. J. 2013, 10, 119. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Wang, Z.; Tan, C.J.; Liao, B.Y.; Zhang, X.; Xu, M.; Dai, Z.; Qiu, S.J.; Huang, X.W.; Sun, J.; et al. Plasma microRNA, a potential biomarker for acute rejection after liver transplantation. Transplantation 2013, 95, 991–999. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.H.; Yi, H.S.; Kim, Y.; Kroh, E.M.; Chien, J.W.; Eaton, K.D.; Goodman, M.T.; Tait, J.F.; Tewari, M.; Pritchard, C.C. Plasma processing conditions substantially influence circulating microRNA biomarker levels. PLoS ONE 2013, 8, e64795. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yeo, J.; Kim, B.; Ha, M.; Kim, V.N. Short structured RNAs with low GC content are selectively lost during extraction from a small number of cells. Mol. Cell 2012, 46, 893–895. [Google Scholar] [CrossRef] [PubMed]
- Whitney, A.R.; Diehn, M.; Popper, S.J.; Alizadeh, A.A.; Boldrick, J.C.; Relman, D.A.; Brown, P.O. Individuality and variation in gene expression patterns in human blood. Proc. Natl. Acad. Sci. USA 2003, 100, 1896–1901. [Google Scholar] [CrossRef] [PubMed]
- Watson, A.K.; Witwer, K.W. Do platform-specific factors explain microRNA profiling disparities? Clin. Chem. 2012, 58, 472–474. [Google Scholar] [CrossRef] [PubMed]
- Dickinson, B.; Zhang, Y.; Petrick, J.S.; Heck, G.; Ivashuta, S.; Marshall, W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013, 31, 965–967. [Google Scholar] [CrossRef] [PubMed]
- Haider, B.A.; Baras, A.S.; McCall, M.N.; Hertel, J.A.; Cornish, T.C.; Halushka, M.K. A critical evaluation of microRNA biomarkers in non-neoplastic disease. PLoS ONE 2014, 9, e89565. [Google Scholar] [CrossRef] [PubMed]
- Xiang, M.; Zeng, Y.; Yang, R.; Xu, H.; Chen, Z.; Zhong, J.; Xie, H.; Xu, Y.; Zeng, X. U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem. Biophys. Res. Commun. 2014, 454, 210–214. [Google Scholar] [CrossRef] [PubMed]
- McDermott, A.M.; Kerin, M.J.; Miller, N. Identification and validation of miRNAs as endogenous controls for RQ-PCR in blood specimens for breast cancer studies. PLoS ONE 2013, 8, e83718. [Google Scholar] [CrossRef] [PubMed]
- Murata, K.; Furu, M.; Yoshitomi, H.; Ishikawa, M.; Shibuya, H.; Hashimoto, M.; Imura, Y.; Fujii, T.; Ito, H.; Mimori, T.; et al. Comprehensive microRNA analysis identifies miR-24 and miR-125a-5p as plasma biomarkers for rheumatoid arthritis. PLoS ONE 2013, 8, e69118. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Circulating miRNA | Cancer Type | Type of Biomarker | Body Fluid Type | Cohort Size | Reference | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| let-7a | Prostate | D | Blood | 75 | 27 | [52] |
| Colorectal | D | Serum exosomes | 88 | 11 | [53] | |
| D | Plasma | 51 | 26 | [54] | ||
| MDS | PG | Plasma | 50 | 76 | [55] | |
| Gastric | D | Plasma | 69 | 30 | [56] | |
| let-7a/b | Lung | D, PG | Serum/plasma | 220 | 220 | [57] |
| let-7b | HCC | D, PG | Serum | 120 | 30 | [58] |
| Ovarian | D | Serum | 18 | 12 | [59] | |
| let-7c | Lung | D | Plasma | 20 | 360 | [60] |
| Breast | D | Serum | 90 | 64 | [61] | |
| let-7c/i/f | Gastric | D | Serum | 214 | 424 | [62] |
| let-7d | Prostate | PG | Plasma | 50 | 10 | [63] |
| let-7e | MM | PG | Serum | 121 | 30 | [64] |
| Thyroid | D | Serum | 95 | 44 | [65] | |
| let-7f | Ovarian | D, PG | Plasma | 360 | 200 | [66] |
| Colorectal | D | Feces | 51 | 26 | [54] | |
| Lung | D, PG | Plasma vesicles | 106 | 68 | [67] | |
| HCC | D, PG | Serum | 90 | 60 | [68] | |
| let-7i | Lung | PG | Serum | 10 | 20 | [69] |
| Ovarian | D | Serum/plasma | 25 | 25 | [70] | |
| miR-10 | Breast | D, PG | Serum | 113 | - | [71] |
| PG | Serum | 89 | 29 | [72] | ||
| PG | Cerebrospinal | 16 | 15 | [73] | ||
| Lung | D | Serum | 42 | 28 | [74] | |
| PG | Cerebrospinal | 28 | 15 | [73] | ||
| Glioblastoma | D | Cerebrospinal | 19 | 15 | [73] | |
| Oesophageal | D | Serum | 50 | 50 | [75] | |
| miR-16 | Oral | D | Serum | 30 | 26 | [76] |
| Prostate | D | Serum | 21 | 15 | [77] | |
| D | Serum | 73 | 20 | [78] | ||
| Breast | D | Serum | 76 | 76 | [79] | |
| Osteosarcoma | D | Serum | 20 | 20 | [80] | |
| Gastric | D, PG | Plasma | 30 | 18 | [81] | |
| D, PG | Serum | 50 | 47 | [82] | ||
| Liver | D | Serum | 90 | 60 | [68] | |
| Oesophageal | D, PG | Plasma | 38 | 19 | [83] | |
| miR-17 | Gastric | D, PG | Serum | 50 | 47 | [82] |
| Liver | D | Serum | 90 | 60 | [68] | |
| Oesophageal | D, PG | Plasma | 38 | 19 | [83] | |
| miR-18a | Oesophageal | D | Serum | 106 | 54 | [84] |
| Gastric | D | Plasma | 104 | 65 | [85] | |
| Breast | D, PD | Serum | 108 | 75 | [86] | |
| Colorectal | D | Stool | 198 | 198 | [87] | |
| miR-17/19a | Melanoma | PD | Plasma | 13 | 13 | [88] |
| miR-19a | Breast | PD | Serum | 30 | 38 | [89] |
| D | Serum | 63 | 21 | [90] | ||
| D, PG | Serum | 113 | - | [71] | ||
| Bladder | D | Plasma | 50 | 50 | [91] | |
| Colorectal | D, PG | Serum | 90 | 12 | [92] | |
| MM | D,PG,PD | Serum | 108 | 56 | [93] | |
| Lung | D, PG | Serum | 201 | 103 | [94] | |
| miR-19b | Gastric | D, PG | Plasma | 30 | 18 | [81] |
| Lung | D, PG | Serum | 94 | 94 | [95] | |
| miR-20a | Prostate | D | Plasma | 82 | - | [96] |
| Lung | D | Plasma | 126 | 60 | [97] | |
| Osteosarcoma | D | Serum | 20 | 20 | [80] | |
| Colorectal | D | Feces | 397 | 198 | [98] | |
| Esophageal | D | Plasma | 70 | 40 | [99] | |
| miR-17/-92 | Colorectal | PD | Serum | 37 | 7 | [100] |
| Colorectal | D | Plasma | 90 | 90 | [101] | |
| miR-92 | Ovarian | D | Serum | 28 | 15 | [102] |
| miR-92a | Colorectal | D | Plasma | 152 | 75 | [103] |
| Leukemia | D | Plasma | 77 | 16 | [104] | |
| miR-92a/b | Prostate | D | Serum | 21 | 15 | [77] |
| miR-106a | Gastric | D, PG | Plasma | 48 | 22 | [105] |
| Colorectal | D | Feces | 117 | 107 | [106] | |
| D | Plasma | 100 | 79 | [107] | ||
| PG | Serum | 175 | 130 | [108] | ||
| miR-17/-106a/b | Gastric | D | Plasma | 69 | 30 | [56] |
| miR-17/-106b | Gastric | D | Serum | 72 | 36 | [109] |
| miR-106b | Liver | D | Plasma | 47 | 61 | [110] |
| D | Serum exosomes | 20 | 40 | [111] | ||
| Breast | D, PG | Plasma | 173 | 50 | [112] | |
| Gastric | D, PG | Plasma | 20 | 20 | [113] | |
| Bladder | D | Urine | 112 | 78 | [114] | |
| Ovarian | D | Serum | 31 | 31 | [115] | |
| miR-21 | DLBCL | D | Serum | 60 | 43 | [11] |
| D,PD,PG | Serum | 112 | 45 | [116] | ||
| D | Serum | 60 | 43 | [11] | ||
| D,PD,PG | Serum | 62 | 50 | [117] | ||
| HL | D | Plasma | 42 | 20 | [118] | |
| CNS lymphoma | D, PG | Serum | 37 | 53 | [119] | |
| Breast | D | Plasma | 14 | 8 | [120] | |
| D, PG | Serum | 62 | 10 | [121] | ||
| D, PG | Serum | 30 | 60 | [122] | ||
| D, PG | Serum | 50 | 82 | [123] | ||
| PG | Serum | 326 | 223 | [124] | ||
| D | Plasma | 114 | 116 | [125] | ||
| D | Urine | 24 | 24 | [126] | ||
| PG | Serum | 113 | - | [71] | ||
| Gastric | D | Plasma | 69 | 30 | [56] | |
| D, PG | Plasma | 42 | - | [127] | ||
| PG | Serum | 103 | - | [128] | ||
| PG | Serum | 79 | - | [129] | ||
| PG | Serum | 64 | 64 | [130] | ||
| PG | Plasma | 69 | - | [131] | ||
| D | Serum | 50 | 50 | [132] | ||
| Glioblastoma | PG | Serum | 30 | 30 | [39] | |
| D | Plasma | 10 | 10 | [133] | ||
| Ovarian | D | Serum | 28 | 15 | [102] | |
| D, PG | Serum | 94 | 40 | [134] | ||
| D | Serum | 60 | 10 | [135] | ||
| Pancreatic | D | Plasma | 49 | 36 | [136] | |
| D, PG | Plasma | 32 | 30 | [137] | ||
| D | Plasma | 24 | 24 | [138] | ||
| D | Stool | 30 | 15 | [139] | ||
| D | Serum | 22 | 14 | [140] | ||
| D | Plasma | 30 | 26 | [141] | ||
| D | Saliva | 7 | 4 | [142] | ||
| Prostate | PD | Plasma | 82 | - | [96] | |
| PD | Serum | 56 | - | [143] | ||
| Colorectal | PD | Serum | 37 | 7 | [100] | |
| D, PG | Serum | 40 | 40 | [144] | ||
| D | Serum | 160 | 77 | [144] | ||
| D, PG | Serum | 186 | 96 | [145] | ||
| D | Serum | 200 | 130 | [146] | ||
| PG | Serum | 102 | - | [147] | ||
| D | Serum | 56 | 197 | [148] | ||
| Melanoma | PD | Plasma | 13 | 13 | [88] | |
| Lung | D, PG | Plasma | 25 | 25 | [97] | |
| D, PG | Serum | 152 | 300 | [149] | ||
| D | BAL + sputum | 21 | 10 | [150] | ||
| D, PG | Serum | 80 | 60 | [151] | ||
| Neck | D | Plasma | 50 | 36 | [152] | |
| Oesophageal | D | Plasma | 50 | 20 | [153] | |
| D | Saliva | 39 | 19 | [154] | ||
| D, PG | Plasma | 38 | 19 | [83] | ||
| Liver | D | Serum | 52 | 43 | [155] | |
| PG | Serum | 224 | - | [156] | ||
| D, PG | Serum | 30 | 60 | [157] | ||
| D | Serum | 23 | 17 | [158] | ||
| Biliary tract | D | Plasma | 94 | 73 | [159] | |
| Nasopharyngeal | D | Plasma | 217 | 73 | [160] | |
| Osteosarcoma | D, PG | Plasma | 40 | 40 | [161] | |
| miR-29a/b/c | Osteosarcoma | D, PG | Serum | 80 | 80 | [162] |
| miR-29a | Colorectal | D | Plasma | 152 | 75 | [103] |
| D | Serum | 38 | 36 | [163] | ||
| D | Serum | 30 | 26 | [164] | ||
| Breast | D | Serum | 63 | 90 | [165] | |
| D | Serum | 20 | 20 | [166] | ||
| Oral | D | Serum | 30 | 26 | [76] | |
| Ovarian | D | Serum | 28 | 15 | [102] | |
| miR-29b | Colorectal | D, PG | Serum | 55 | 55 | [167] |
| miR-29c | Colorectal | PG | Serum | 103 | 37 | [168] |
| Lung | D | Serum | 70 | 48 | [169] | |
| Nasopharyngeal | D | Serum | 160 | 143 | [170] | |
| miR-30a | Lung | D | Plasma | 60 | 75 | [171] |
| Esophageal | D | Serum exosomes | 18 | 29 | [172] | |
| miR-30c | Prostate | D | Plasma | 105 | 115 | [173] |
| D | Plasma | 59 | 27 | [174] | ||
| Lung | D | Serum | 80 | 40 | [175] | |
| miR-30d | Lung | PG | Serum | 82 | 50 | [176] |
| PG | Serum | 303 | - | [177] | ||
| miR-30e | Liver | D | Serum | 39 | 31 | [178] |
| miR-34a | Breast | D, PG | Serum | 89 | 29 | [74] |
| Lung | D | Blood | 22 | 27 | [179] | |
| miR-34b | Prostate | D | Serum | 21 | 15 | [77] |
| Osteosarcoma | D | Plasma | 133 | 133 | [180] | |
| miR-34b/c | Breast | D | Serum | 15 | 15 | [181] |
| miR-34c | Lung | D | Serum | 17 | 19 | [182] |
| miR-125a | Oral | D | Saliva | 50 | 62 | [183] |
| Lung | D | Serum | 70 | 70 | [184] | |
| Breast | PG | Serum | 300 | - | [185] | |
| Liver | PG | Serum | 120 | 255 | [186] | |
| miR-125b | Breast | PD | Serum | 56 | 10 | [187] |
| D | Plasma | 197 | 142 | [125] | ||
| Lung | D, PG | Serum | 193 | 110 | [188] | |
| PG, PD | Serum | 260 | 260 | [189] | ||
| Oral | D | Plasma | 85 | 46 | [190] | |
| Colorectal | D | Serum | 160 | 77 | [191] | |
| Glioma | D | Serum | 33 | 33 | [192] | |
| Melanoma | D | Serum exosomes | 21 | 35 | [193] | |
| miR-155 | Breast | D | Serum | 63 | 21 | [90] |
| D | Plasma/serum | 184 | 75 | [194] | ||
| D | Serum | 20 | 10 | [195] | ||
| D | Serum | 103 | 55 | [196] | ||
| D, PG | Serum | 89 | 29 | [74] | ||
| PG | Serum | 32 | 120 | [197] | ||
| Colorectal | D, PG | Serum | 146 | 60 | [198] | |
| Lung | D | Serum | 36 | 32 | [199] | |
| D | Serum/plasma | 220 | 220 | [57] | ||
| Esophageal | D, PG | Plasma | 60 | 60 | [200] | |
| AML | D | Serum | 140 | 135 | [201] | |
| DLBCL | D | Serum | 75 | 77 | [202] | |
| D | Serum | 60 | 43 | [11] | ||
| CLL | PG, PD | Plasma | 228 | - | [203] | |
| ATL | PG | Plasma | 35 | - | [204] | |
| miR-200a/b/c/141 | Ovarian | D | Serum exosomes | 50 | 20 | [135] |
| miR-200a/b/c | Ovarian | D, PG | Serum | 70 | 70 | [205] |
| D | Serum | 28 | 28 | [206] | ||
| miR-200c/141 | Breast | D, PG | Blood | 57 | 20 | [207] |
| miR-200c/141 | Ovarian | D, PG | Serum | 93 | 50 | [208] |
| miR-200a | Oral | D | Saliva | 50 | 62 | [183] |
| miR-200b | Prostate | PG, PD | Serum/plasma | 97 | - | [209] |
| miR-200c | Colorectal | D | Plasma | 78 | 86 | [210] |
| PG | Serum | 206 | 24 | [211] | ||
| Gastric | D, PG | Serum | 98 | 100 | [212] | |
| D, PG | Blood | 52 | 15 | [213] | ||
| Lung | D, PG | Serum | 70 | 44 | [214] | |
| Esophageal | PG, PD | Serum | 64 | - | [215] | |
| miR-141 | Urinary tract | D | Serum | 44 | 34 | [216] |
| Lung | D | Serum | 42 | 28 | [74] | |
| Prostate | D | Serum | 25 | 25 | [12] | |
| PG | Serum | 113 | - | [217] | ||
| D | Serum | 21 | 15 | [77] | ||
| D | Plasma vesicles | 78 | 28 | [218] | ||
| D | Serum exosomes | 71 | 80 | [219] | ||
| PG | Serum | 30 | 26 | [220] | ||
| Colorectal | PG | Plasma | 185 | 76 | [221] | |
| miR-429 | Lung | D, PG | Serum | 70 | 48 | [169] |
| miR-210 | DLBCL | D | Serum | 60 | 43 | [11] |
| Pancreatic | D | Plasma | 49 | 36 | [136] | |
| D | Plasma | 22 | 25 | [222] | ||
| Renal | D | Serum | 78 | 42 | [223] | |
| D | Serum | 34 | 23 | [224] | ||
| Breast | PG, PD | Plasma | 69 | 43 | [225] | |
| Pancreatic | D | Saliva | 7 | 4 | [142] | |
| D, PG | Pancreatic juice | 6 | 6 | [226] | ||
| D | Plasma | 30 | 26 | [141] | ||
| Bladder | D, PG | Serum | 168 | 104 | [227] | |
| D | Urine | 94 | 56 | [228] | ||
| Glioma | D, PG | Serum | 136 | 50 | [229] | |
| Liver | PD, PG | Serum | 113 | 39 | [230] | |
| miR-221 | Colorectal | D | Plasma | 103 | 37 | [231] |
| D | Stool | 198 | 198 | [87] | ||
| Prostate | PG | Plasma | 82 | - | [96] | |
| Leukemia | D | Plasma | 79 | 37 | [232] | |
| Liver | D | Serum | 20 | 40 | [111] | |
| Larynx | D | Plasma | 30 | 30 | [233] | |
| Glioma | D, PG | Plasma | 50 | 51 | [234] | |
| Melanoma | D, PG | Serum | 72 | 54 | [235] | |
| Renal | PG | Plasma | 77 | - | [236] | |
| miR-375 | Breast | PG | Serum | 68 | - | [237] |
| Lung | D, PG | Plasma | 217 | 217 | [238] | |
| PG | Serum | 113 | - | [217] | ||
| Prostate | PG | Serum | 47 | 72 | [218] | |
| PG | Serum | 84 | - | [239] | ||
| PG | Plasma | 100 | - | [240] | ||
| D | Plasma | 78 | 28 | [218] | ||
| Oesophageal | D | Plasma | 38 | 19 | [83] | |
| PG | Serum | 194 | 94 | [241] | ||
| D | Plasma | 50 | 20 | [153] | ||
| Liver | D | Serum | 78 | 156 | [242] | |
| Colorectal | D | Plasma | 88 | 40 | [243] | |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. Int. J. Mol. Sci. 2016, 17, 627. https://doi.org/10.3390/ijms17050627
Larrea E, Sole C, Manterola L, Goicoechea I, Armesto M, Arestin M, Caffarel MM, Araujo AM, Araiz M, Fernandez-Mercado M, et al. New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. International Journal of Molecular Sciences. 2016; 17(5):627. https://doi.org/10.3390/ijms17050627
Chicago/Turabian StyleLarrea, Erika, Carla Sole, Lorea Manterola, Ibai Goicoechea, María Armesto, María Arestin, María M. Caffarel, Angela M. Araujo, María Araiz, Marta Fernandez-Mercado, and et al. 2016. "New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies" International Journal of Molecular Sciences 17, no. 5: 627. https://doi.org/10.3390/ijms17050627
APA StyleLarrea, E., Sole, C., Manterola, L., Goicoechea, I., Armesto, M., Arestin, M., Caffarel, M. M., Araujo, A. M., Araiz, M., Fernandez-Mercado, M., & Lawrie, C. H. (2016). New Concepts in Cancer Biomarkers: Circulating miRNAs in Liquid Biopsies. International Journal of Molecular Sciences, 17(5), 627. https://doi.org/10.3390/ijms17050627