Global Metabolic Regulation of the Snow Alga Chlamydomonas nivalis in Response to Nitrate or Phosphate Deprivation by a Metabolome Profile Analysis
Abstract
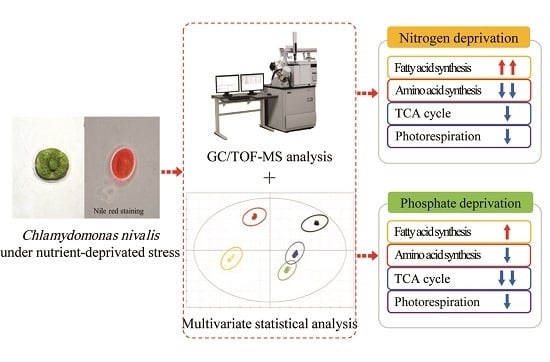
:1. Introduction
2. Results and Discussion
2.1. Identification and Overview of the Metabolites
2.2. Discrimination of Metabolomic Data by OPLS-DA (the Orthogonal Partial Least Squares Discriminant Analysis)
2.3. Selection of the Responding Biomarkers
2.4. Abundance of the Responding Biomarkers in the Different Groups
2.4.1. Responding Biomarkers in the Nitrate-Deprived Group
2.4.2. Responding Biomarkers in the Phosphate-Deprived Group
2.5. Metabolic Pathway and Regulation Analysis under Stresses
3. Experimental Section
3.1. Materials
3.2. Algal Culture, Stress Treatment and Biomass Harvest
3.3. Extraction and Derivatization of the Cellular Compounds
3.4. Metabolome Profile Analyses by Gas Chromatography Time-of-Flight Mass Spectrometry (GC/TOF-MS)
3.4.1. Gas Chromatography Procedure
3.4.2. Mass Spectrometry
3.4.3. Data Processing and Normalization
3.5. Multivariate Statistical Analyses
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wiencke, C.; Clayton, M.N. Biology of polar benthic algae. Bot. Mar. 2009, 52, 479–481. [Google Scholar] [CrossRef]
- Schwarz, R.; Forchhammer, K. Acclimation of unicellular cyanobacteria to macronutrient deficiency: Emergence of a complex network of cellular responses. Microbiol. SGM 2005, 151, 2503–2514. [Google Scholar] [CrossRef] [PubMed]
- Gaude, N.; Brehelin, C.; Tischendorf, G.; Kessler, F.; Doermann, P. Nitrogen deficiency in arabidopsis affects galactolipid composition and gene expression and results in accumulation of fatty acid phytyl esters. Plant J. 2007, 49, 729–739. [Google Scholar] [CrossRef] [PubMed]
- El-Sheek, M.M.; Rady, A.A. Effect of phosphorus starvation on growth, photosynthesis and some metabolic processes in the unicellular green alga chlorella kessleri. Phyton 1995, 35, 139–151. [Google Scholar]
- Lu, N.; Wei, D.; Chen, F.; Yang, S.-T. Lipidomic profiling reveals lipid regulation in the snow alga Chlamydomonas nivalis in response to nitrate or phosphate deprivation. Process Biochem. 2013, 48, 605–613. [Google Scholar] [CrossRef]
- Schmollinger, S.; Muehlhaus, T.; Boyle, N.R.; Blaby, I.K.; Casero, D.; Mettler, T.; Moseley, J.L.; Kropat, J.; Sommer, F.; Strenkert, D.; et al. Nitrogen-sparing mechanisms in Chlamydomonas affect the transcriptome, the proteome, and photosynthetic metabolism. Plant Cell 2014, 26, 1410–1435. [Google Scholar] [CrossRef] [PubMed]
- Soga, T.; Ohashi, Y.; Ueno, Y.; Naraoka, H.; Tomita, M.; Nishioka, T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003, 2, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-J.; Wang, H.; Gargouri, M.; Deshpande, R.R.; Skepper, J.N.; Holguin, F.O.; Juergens, M.T.; Shachar-Hill, Y.; Hicks, L.M.; Gang, D.R. The response of Chlamydomonas reinhardtii to nitrogen deprivation: A systems biology analysis. Plant J. 2015, 81, 611–624. [Google Scholar] [CrossRef] [PubMed]
- Feng, T.-Y.; Yang, Z.-K.; Zheng, J.-W.; Xie, Y.; Li, D.-W.; Murugan, S.B.; Yang, W.-D.; Liu, J.-S.; Li, H.-Y. Examination of metabolic responses to phosphorus limitation via proteomic analyses in the marine diatom phaeodactylum tricornutum. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Bielecka, M.; Watanabe, M.; Morcuende, R.; Scheible, W.-R.; Hawkesford, M.J.; Hesse, H.; Hoefgen, R. Transcriptome and metabolome analysis of plant sulfate starvation and resupply provides novel information on transcriptional regulation of metabolism associated with sulfur, nitrogen and phosphorus nutritional responses in arabidopsis. Front. Plant Sci. 2015, 5, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Beisken, S.; Eiden, M.; Salek, R.M. Getting the right answers: Understanding metabolomics challenges. Expert Rev. Mol. Diagn. 2015, 15, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Peterson, A.C.; Balloon, A.J.; Westphall, M.S.; Coon, J.J. Development of a GC/quadrupole-orbitrap mass spectrometer, part II: New approaches for discovery metabolomics. Anal. Chem. 2014, 86, 10044–10051. [Google Scholar] [CrossRef] [PubMed]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell. Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [PubMed]
- Veyel, D.; Erban, A.; Fehrle, I.; Kopka, J.; Schroda, M. Rationales and approaches for studying metabolism in eukaryotic microalgae. Metabolites 2014, 4, 184–217. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Fiehn, O. High quality metabolomic data for Chlamydomonas reinhardtii. Plant Methods 2008, 4. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, S.; Johansson, E.; Sjostrom, L.; Mellerowicz, E.J.; Edlund, U.; Shockcor, J.P.; Gottfries, J.; Moritz, T.; Trygg, J. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using opls class models. Anal. Chem. 2008, 80, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Cajka, T.; Fiehn, O. Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics. Anal. Chem. 2016, 88, 524–545. [Google Scholar] [CrossRef] [PubMed]
- Wienkoop, S.; Weiss, J.; May, P.; Kempa, S.; Irgang, S.; Recuenco-Munoz, L.; Pietzke, M.; Schwemmer, T.; Rupprecht, J.; Egelhofer, V.; et al. Targeted proteomics for Chlamydomonas reinhardtii combined with rapid subcellular protein fractionation, metabolomics and metabolic flux analyses. Mol. Biosyst. 2010, 6, 1018–1031. [Google Scholar] [CrossRef] [PubMed]
- Diaz, G.; Melis, M.; Batetta, B.; Angius, F.; Falchi, A.M. Hydrophobic characterization of intracellular lipids in situ by nile red red/yellow emission ratio. Micron 2008, 39, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Fernie, A.R.; Obata, T.; Allen, A.E.; Araujo, W.L.; Bowler, C. Leveraging metabolomics for functional investigations in sequenced marine diatoms. Trends Plant Sci. 2012, 17, 395–403. [Google Scholar] [CrossRef] [PubMed]
- Vidoudez, C.; Pohnert, G. Comparative metabolomics of the diatom skeletonema marinoi in different growth phases. Metabolomics 2012, 8, 654–669. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward pathway engineering: A new database of genetic and molecular pathways. Sci. Technol. Jpn. 1996, 59, 34–38. [Google Scholar]
- Pandhal, J.; Noirel, J.; Wright, P.C.; Biggs, C.A. A systems biology approach to investigate the response of Synechocystis sp. PCC6803 to a high salt environment. Saline Syst. 2009, 5. [Google Scholar] [CrossRef] [PubMed]
- Hockin, N.L.; Mock, T.; Mulholland, F.; Kopriva, S.; Malin, G. The response of diatom central carbon metabolism to nitrogen starvation is different from that of green algae and higher plants. Plant Physiol. 2012, 158, 299–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, N.; Wei, D.; Jiang, X.-L.; Chen, F.; Yang, S.-T. Regulation of lipid metabolism in the snow alga Chlamydomonas nivalis in response to NACL stress: An integrated analysis by cytomic and lipidomic approaches. Process Biochem. 2012, 47, 1163–1170. [Google Scholar] [CrossRef]
- Lu, N.; Wei, D.; Jiang, X.-L.; Chen, F.; Yang, S.-T. Fatty acids profiling and biomarker identification in snow alga Chlamydomonas nivalis by NACL stress using GC/MS and multivariate statistical analysis. Anal. Lett. 2012, 45, 1172–1183. [Google Scholar] [CrossRef]
- Lu, N.; Wei, D.; Chen, F.; Yang, S.-T. Lipidomic profiling and discovery of lipid biomarkers in snow alga Chlamydomonas nivalis under salt stress. Eur. J. Lipid Sci. Technol. 2012, 114, 253–265. [Google Scholar] [CrossRef]
- Guerrini, F.; Cangini, M.; Boni, L.; Trost, P.; Pistocchi, R. Metabolic responses of the diatom Achnanthes brevipes (bacillariophyceae) to nutrient limitation. J. Phycol. 2000, 36, 882–890. [Google Scholar] [CrossRef]
- Lai, J.; Yu, Z.; Song, X.; Cao, X.; Han, X. Responses of the growth and biochemical composition of prorocentrum donghaiense to different nitrogen and phosphorus concentrations. J. Exp. Mar. Biol. Ecol. 2011, 405, 6–17. [Google Scholar] [CrossRef]
- Kang, J.; Choi, M.-Y.; Kang, S.; Kwon, H.N.; Wen, H.; Lee, C.H.; Park, M.; Wiklund, S.; Kim, H.J.; Kwon, S.W.; et al. Application of a 1H nuclear magnetic resonance (NMR) metabolomics approach combined with orthogonal projections to latent structure-discriminant analysis as an efficient tool for discriminating between korean and chinese herbal medicines. J. Agric. Food Chem. 2008, 56, 11589–11595. [Google Scholar] [CrossRef] [PubMed]
- Van de Wiel, C.C.M.; van der Linden, C.G.; Scholten, O.E. Improving phosphorus use efficiency in agriculture: Opportunities for breeding. Euphytica 2016, 207, 1–22. [Google Scholar] [CrossRef]
- Blatti, J.L.; Michaud, J.; Burkart, M.D. Engineering fatty acid biosynthesis in microalgae for sustainable biodiesel. Curr. Opin. Chem. Biol. 2013, 17, 496–505. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.T.; Ullrich, N.; Joo, S.; Waffenschmidt, S.; Goodenough, U. Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 2009, 8, 1856–1868. [Google Scholar] [CrossRef] [PubMed]
- Miller, R.; Wu, G.; Deshpande, R.R.; Vieler, A.; Gaertner, K.; Li, X.; Moellering, E.R.; Zaeuner, S.; Cornish, A.J.; Liu, B.; et al. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 2010, 154, 1737–1752. [Google Scholar] [CrossRef] [PubMed]
- Flynn, K. Algal carbon-nitrogen metabolism: A biochemical basis for modelling the interactions between nitrate and ammonium uptake. J. Plankton Res. 1991, 13, 373–387. [Google Scholar] [CrossRef]
- Ito, T.; Tanaka, M.; Shinkawa, H.; Nakada, T.; Ano, Y.; Kurano, N.; Soga, T.; Tomita, M. Metabolic and morphological changes of an oil accumulating trebouxiophycean alga in nitrogen-deficient conditions. Metabolomics 2013, 9, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Theodorou, M.E.; Elrifi, I.R.; Turpin, D.H.; Plaxton, W.C. Effects of phosphorus limitation on respiratory metabolism in the green alga selenastrum minutum. Plant Physiol. 1991, 95, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- Juneja, A.; Ceballos, R.M.; Murthy, G.S. Effects of environmental factors and nutrient availability on the biochemical composition of algae for biofuels production: A review. Energies 2013, 6, 4607–4638. [Google Scholar] [CrossRef]
- Siderius, M.; Musgrave, A.; Ende, H.; Koerten, H.; Cambier, P.; Meer, P. Chlamydomonas eugametos (Chlorophyta) stores phosphate in polyphosphate bodies together with calcium. J. Phycol. 1996, 32, 402–409. [Google Scholar] [CrossRef]
- Hebeler, M.; Hentrich, S.; Mayer, A.; Leibfritz, D.; Grimme, L. Phosphate regulation and compartmentation in Chlamydomonas reinhardtii studied by in vivo p-31-NMR. Photosynth. Res. 1992, 34, 199. [Google Scholar]
- Loera-Quezada, M.M.; Leyva-Gonzalez, M.A.; Lopez-Arredondo, D.; Herrera-Estrella, L. Phosphite cannot be used as a phosphorus source but is non-toxic for microalgae. Plant Sci. 2015, 231, 124–130. [Google Scholar] [CrossRef] [PubMed]
- Muratsubaki, H. Regulation of reductive production of succinate under anaerobic conditions in baker's yeast. J. Biochem. 1987, 102, 705–714. [Google Scholar] [PubMed]
- Sirithanakorn, C.; Adina-Zada, A.; Wallace, J.C.; Jitrapakdee, S.; Attwood, P.V. Mechanisms of inhibition of Rhizobium etli pyruvate carboxylase by l-aspartate. Biochemistry 2014, 53, 7100–7106. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Sato, Y.; Furumichi, M.; Tanabe, M. Kegg for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012, 40, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Lukes, M.; Prochazkova, L.; Shmidt, V.; Nedbalova, L.; Kaftan, D. Temperature dependence of photosynthesis and thylakoid lipid composition in the red snow alga Chlamydomonas cf. nivalis (Chlorophyceae). FEMS Microbiol. Ecol. 2014, 89, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Irihimovitch, V.; Yehudai-Resheff, S. Phosphate and sulfur limitation responses in the chloroplast of Chlamydomonas reinhardtii. FEMS Microbiol. Lett. 2008, 283, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Bolling, C.; Fiehn, O. Metabolite profiling of Chlamydomonas reinhardtii under nutrient deprivation. Plant Physiol. 2005, 139, 1995–2005. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O.; Kind, T. Metabolite profiling in blood plasma. Methods Mol. Biol. 2007, 358, 3–17. [Google Scholar] [PubMed]
- Kind, T.; Wohlgemuth, G.; Lee, D.Y.; Lu, Y.; Palazoglu, M.; Shahbaz, S.; Fiehn, O. Fiehnlib: Mass spectral and retention index libraries for metabolomics based on quadrupole and time-of-fight gas chromatography/mass spectrometry. Anal. Chem. 2009, 81, 10038–10048. [Google Scholar] [CrossRef] [PubMed]







| Number | Identified Metabolites | VIP | p[1] | p(corr)[1] | Change Rate in Abundance, % (N Deprivation vs. the Control) |
|---|---|---|---|---|---|
| 1 | Linolenic acid | 5.48 | 0.22 | 0.65 | 5.13 |
| 2 | α-Ketoglutaric acid | 3.24 | 0.28 | 0.94 | −55.11 |
| 3 | Stearic acid | 2.60 | −0.20 | −0.88 | 67.15 |
| 4 | Putrescine | 2.48 | 0.24 | 0.97 | −29.31 |
| 5 | Glutamine | 2.42 | 0.22 | 0.95 | −21.44 |
| 6 | Asparagine | 2.33 | 0.20 | 0.93 | −58.90 |
| 7 | Palmitic acid | 2.01 | −0.20 | −0.97 | 67.00 |
| 8 | Uracil | 1.92 | 0.20 | 0.99 | −11.27 |
| 9 | Adenosine | 1.87 | 0.16 | 0.93 | −28.98 |
| 10 | Glutamic acid | 1.86 | 0.18 | 0.97 | −35.24 |
| 11 | Nicotinamide | 1.75 | 0.19 | 0.99 | −8.17 |
| 12 | Adenine | 1.74 | 0.17 | 0.96 | −30.60 |
| 13 | Succinic acid | 1.71 | 0.17 | 0.94 | 8.36 |
| 14 | Valine | 1.65 | 0.17 | 0.99 | −6.68 |
| 15 | Isoleucine | 1.61 | −0.10 | −0.80 | 75.65 |
| 16 | Trisaccharide | 1.59 | 0.17 | 1.00 | −2.16 |
| 17 | Citric acid | 1.52 | 0.13 | 0.91 | −33.14 |
| 18 | Pentadecanoic acid | 1.45 | −0.13 | −0.95 | 75.54 |
| 19 | Alanine | 1.43 | 0.15 | 0.98 | −56.06 |
| 20 | Malic acid | 1.41 | 0.15 | 0.99 | −11.30 |
| 21 | Fumaric acid | 1.36 | 0.13 | 0.94 | −34.74 |
| 22 | Aspartic acid | 1.35 | 0.11 | 0.92 | −5.22 |
| 23 | Proline | 1.34 | 0.13 | 0.97 | −1.03 |
| 24 | Serine | 1.30 | 0.11 | 0.93 | −9.06 |
| 25 | Phenylalanine | 1.19 | 0.12 | 0.97 | −29.42 |
| 26 | Histidine | 1.13 | −0.12 | −0.99 | 86.94 |
| 27 | Tryptophan | 1.11 | −0.12 | −1.00 | 85.84 |
| 28 | Shikimic acid | 1.11 | 0.12 | 1.00 | −35.76 |
| 29 | Leucine | 1.09 | 0.11 | 0.97 | −27.64 |
| 30 | Lysine | 1.01 | 0.11 | 0.99 | −4.38 |
| Number | Identified Metabolites | VIP | p[1] | p(corr)[1] | Change Rate in Abundance, % (P deprivation vs. the Control) |
|---|---|---|---|---|---|
| 1 | Linolenic acid | 3.42 | 0.30 | 0.98 | −22.77 |
| 2 | Stearic acid | 2.89 | −0.25 | −0.96 | 47.74 |
| 3 | Trisaccharide | 2.75 | −0.12 | −0.88 | 24.77 |
| 4 | Adenosine | 2.53 | 0.21 | 0.98 | −43.47 |
| 5 | Glutamic acid | 2.49 | 0.21 | 0.97 | 10.29 |
| 6 | α-Ketoglutaric acid | 2.34 | 0.25 | 0.99 | −22.15 |
| 7 | Putrescine | 2.16 | 0.24 | 1.00 | −19.51 |
| 8 | Fumaric acid | 1.84 | 0.17 | 0.99 | −55.15 |
| 9 | Trehalose | 1.81 | 0.10 | 0.93 | −20.32 |
| 10 | Threonic acid | 1.79 | 0.11 | 0.92 | 1.05 |
| 11 | Shikimic acid | 1.75 | 0.14 | 0.97 | −42.50 |
| 12 | Nicotinamide | 1.63 | 0.15 | 0.94 | −3.00 |
| 13 | Valine | 1.60 | 0.15 | 0.98 | −4.15 |
| 14 | Succinic acid | 1.51 | 0.16 | 0.94 | −1.38 |
| 15 | Proline | 1.49 | 0.11 | 0.95 | −1.51 |
| 16 | Aspartic acid | 1.49 | −0.13 | −0.97 | 42.85 |
| 17 | Benzoic acid | 1.48 | 0.05 | 0.64 | 6.81 |
| 18 | Histidine | 1.46 | 0.13 | 0.97 | −23.75 |
| 19 | Uracil | 1.42 | 0.15 | 0.99 | −1.08 |
| 20 | Sucrose | 1.41 | 0.04 | 0.73 | 9.17 |
| 21 | Malic acid | 1.39 | 0.10 | 0.89 | −0.39 |
| 22 | Serine | 1.37 | 0.10 | 0.93 | −5.55 |
| 23 | Threitol | 1.36 | 0.05 | 0.71 | −1.11 |
| 24 | Phenylalanine | 1.35 | 0.15 | 0.99 | −39.56 |
| 25 | Erythronic acid lactone | 1.30 | 0.08 | 0.80 | 1.74 |
| 26 | Leucine | 1.29 | 0.13 | 0.99 | −38.43 |
| 27 | Adenosine-5-phosphate | 1.27 | 0.12 | 0.97 | −4.98 |
| 28 | Palmitic acid | 1.23 | −0.13 | −0.79 | 25.12 |
| 29 | 3-Phosphoglycerate | 1.22 | −0.12 | −0.99 | 297.76 |
| 30 | Fructose-6-phosphate | 1.21 | 0.13 | 0.99 | −21.05 |
| 31 | Inositol-4-monophosphate | 1.21 | 0.12 | 0.99 | −37.26 |
| 32 | Alanine | 1.19 | 0.13 | 0.99 | −27.64 |
| 33 | Pyrophosphate | 1.13 | −0.12 | −0.99 | 41.90 |
| 34 | Gluconic acid | 1.09 | 0.08 | 0.93 | −35.19 |
| 35 | Asparagine | 1.09 | 0.12 | 0.99 | −3.23 |
| 36 | Pentadecanoic acid | 1.07 | −0.10 | −0.96 | 32.98 |
| 37 | Tyrosine | 1.02 | 0.10 | 0.97 | 1.73 |
| 38 | Threonine | 1.00 | −0.08 | −0.94 | 23.50 |
| 39 | Cellobiose | 1.00 | 0.10 | 0.94 | 8.64 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, N.; Chen, J.-H.; Wei, D.; Chen, F.; Chen, G. Global Metabolic Regulation of the Snow Alga Chlamydomonas nivalis in Response to Nitrate or Phosphate Deprivation by a Metabolome Profile Analysis. Int. J. Mol. Sci. 2016, 17, 694. https://doi.org/10.3390/ijms17050694
Lu N, Chen J-H, Wei D, Chen F, Chen G. Global Metabolic Regulation of the Snow Alga Chlamydomonas nivalis in Response to Nitrate or Phosphate Deprivation by a Metabolome Profile Analysis. International Journal of Molecular Sciences. 2016; 17(5):694. https://doi.org/10.3390/ijms17050694
Chicago/Turabian StyleLu, Na, Jun-Hui Chen, Dong Wei, Feng Chen, and Gu Chen. 2016. "Global Metabolic Regulation of the Snow Alga Chlamydomonas nivalis in Response to Nitrate or Phosphate Deprivation by a Metabolome Profile Analysis" International Journal of Molecular Sciences 17, no. 5: 694. https://doi.org/10.3390/ijms17050694
APA StyleLu, N., Chen, J. -H., Wei, D., Chen, F., & Chen, G. (2016). Global Metabolic Regulation of the Snow Alga Chlamydomonas nivalis in Response to Nitrate or Phosphate Deprivation by a Metabolome Profile Analysis. International Journal of Molecular Sciences, 17(5), 694. https://doi.org/10.3390/ijms17050694