Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis
Abstract
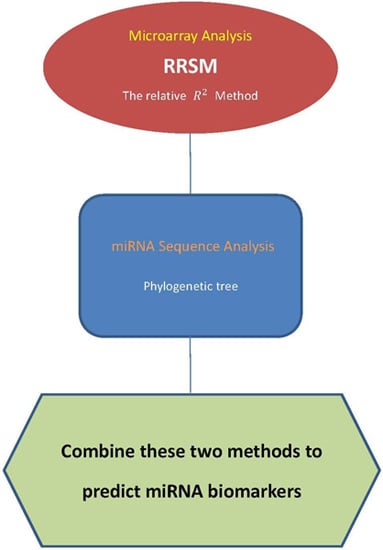
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Microarray Approach
4.2. Phylogenetic Tree
4.3. Procedure of Discovering High-Confidence miRNA Biomarkers
- Step 1.
- Use the stem-loop sequences of miRNAs to build a phylogenetic tree.
- Step 2.
- Use RRSM or other microarray data analysis to select cancer-related miRNA. Cluster miRNAs into different groups subject to the cancer target prediction result; see Table 3.
- Step 3.
- Collect miRNAs in the same clade in the phylogenetic tree of Step 1. If miRNAs in the same clade belong to the same group of miRNAs that are clustered in Step 2, these miRNAs are selected to be high-confidence miRNA biomarkers for particular cancers.
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microrna targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The c-elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Sassen, S.; Miska, E.A.; Caldas, C. Microrna: Implications for cancer. Virchows Arch. 2008, 452, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Foldes-Papp, Z.; Konig, K.; Studier, H.; Buckle, R.; Breunig, H.G.; Uchugonova, A.; Kostner, G.M. Trafficking of mature miRNA-122 into the nucleus of live liver cells. Curr. Pharm. Biotechnol. 2009, 10, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Croce, C.M. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009, 10, 704–714. [Google Scholar] [CrossRef] [PubMed]
- Van Schooneveld, E.; Wouters, M.C.A.; van der Auwera, I.; Peeters, D.J.; Wildiers, H.; van Dam, P.A.; Vergote, I.; Vermeulen, P.B.; Dirix, L.Y.; van Laere, S.J. Expression profiling of cancerous and normal breast tissues identifies micrornas that are differentially expressed in serum from patients with (metastatic) breast cancer and healthy volunteers. Breast Cancer Res. 2012, 14, R34. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.J.; Lin, F.M.; Huang, H.D.; Wang, H. Investigating microrna-target interaction-supported tissues in human cancer tissues based on miRNA and target gene expression profiling. PLoS ONE 2014, 9, e95697. [Google Scholar] [CrossRef] [PubMed]
- Wang, H. Predicting cancer-related miRNAs using expression profiles in tumor tissue. Curr. Pharm. Biotechnol. 2014, 15, 438–444. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.S.; Wang, H. Variance estimation for nucleotide substitution models. Mol. Phylogenet. Evol. 2015, 93, 380–380. [Google Scholar] [CrossRef]
- Wang, H. Confidence intervals for the substitution number in the nucleotide substitution models. Mol. Phylogenet. Evol. 2011, 60, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Hung, S.L. Phylogenetic tree selection by the adjusted k-means approach. J. Appl. Stat. 2012, 39, 643–655. [Google Scholar] [CrossRef]
- Graur, D.; Li, W.-H. Fundamentals of Molecular Evolution, 2nd ed.; Sinauer Associates: Sunderland, MA, USA, 2000; p. 481. [Google Scholar]
- Huang, J.C.; Babak, T.; Corson, T.W.; Chua, G.; Khan, S.; Gallie, B.L.; Hughes, T.R.; Blencowe, B.J.; Frey, B.J.; Morris, Q.D. Using expression profiling data to identify human microRNA targets. Nat. Methods 2007, 4, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, W.H. Increasing microRNA target prediction confidence by the relative R2 method. J. Theor. Biol. 2009, 259, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.J.; Wang, H. Human microRNA target identification by RRSM. J. Theor. Biol. 2011, 286, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, W.J.; Wang, H. RRSM with a data-dependent threshold for miRNA target prediction. J. Theor. Biol. 2013, 337, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011, 39, D152–D157. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. Mirbase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.J.; Zhang, X.; Yang, Y.M.; Li, Z.W.; Du, L.T.; Dong, Z.G.; Qu, A.L.; Jiang, X.M.; Li, P.L.; Wang, C.X. Urinary cell-free microRNA-106b as a novel biomarker for detection of bladder cancer. Med. Oncol. 2014, 31, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Seki, N.; Itesako, T.; Chiyomaru, T.; Nakagawa, M.; Enokida, H. Aberrant expression of microRNAs in bladder cancer. Nat. Rev. Urol. 2013, 10, 396–404. [Google Scholar] [CrossRef] [PubMed]
- Mengual, L.; Lozano, J.J.; Ingelmo-Torres, M.; Gazquez, C.; Ribal, M.J.; Alcaraz, A. Using microRNA profiling in urine samples to develop a non-invasive test for bladder cancer. Int. J. Cancer 2013, 133, 2631–2641. [Google Scholar] [PubMed]
- Adam, L.; Wszolek, M.F.; Liu, C.G.; Jing, W.; Diao, L.X.; Zien, A.; Zhang, J.T.D.; Jackson, D.; Dinney, C.P.N. Plasma microRNA profiles for bladder cancer detection. Urol. Oncol. Semin. Orig. 2013, 31, 1701–1708. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Wang, Z.Z.; Xue, J. Artesunate induces apoptosis of bladder cancer cells by miR-16 regulation of COX-2 expression. Int. J. Mol. Sci. 2014, 15, 14298–14312. [Google Scholar] [CrossRef] [PubMed]
- Itesako, T.; Seki, N.; Yoshino, H.; Chiyomaru, T.; Yamasaki, T.; Hidaka, H.; Yonezawa, T.; Nohata, N.; Kinoshita, T.; Nakagawa, M.; et al. The microrna expression signature of bladder cancer by deep sequencing: The functional significance of the miR-195/497 cluster. PLoS ONE 2014, 9, e84311. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.; Rodriguez, C.E.; Donald, G.W.; Hertzer, K.M.; Jung, X.S.; Chang, H.H.; Moro, A.; Reber, H.A.; Hines, O.J.; Eibl, G. miR-143 decreases cox-2 mRNA stability and expression in pancreatic cancer cells. Biochem. Biophys. Res. Commun. 2013, 439, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Ou, Y.; Wu, K.; Chen, Y.; Sun, W. miR-143 inhibits the metastasis of pancreatic cancer and an associated signaling pathway. Tumour Biol. 2012, 33, 1863–1870. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Fang, S.R.; Gao, L.H.; Tan, Y.; Yang, Z.H. Clinic significance of microRNA-99a expression in human lung adenocarcinoma. J. Surg. Oncol. 2013, 108, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.; Wang, R.; Chen, L.B. miR-100 resensitizes docetaxel-resistant human lung adenocarcinoma cells (SPC-A1) to docetaxel by targeting Plk1. Cancer Lett. 2012, 317, 184–191. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.L.; Rigoutsos, I. miR-103a-3p targets the 5′ UTR of GPRC5A in pancreatic cells. RNA 2014, 20, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhang, L.; Yin, Z.Y.; Fan, X.L.; Hu, B.; Wang, L.Q.; Zhang, D. miR-107 regulates cisplatin chemosensitivity of a549 non small cell lung cancer cell line by targeting cyclin dependent kinase 8. Int. J. Clin. Exp. Pathol. 2014, 7, 7236–7241. [Google Scholar] [PubMed]
- Iliopoulos, D.; Rotem, A.; Struhl, K. Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res. 2011, 71, 5144–5153. [Google Scholar] [CrossRef] [PubMed]
- Leivonen, S.K.; Rokka, A.; Ostling, P.; Kohonen, P.; Corthals, G.L.; Kallioniemi, O.; Perala, M. Identification of miR-193b targets in breast cancer cells and systems biological analysis of their functional impact. Mol. Cell. Proteom. 2011, 10. [Google Scholar] [CrossRef] [PubMed]
- Song, M.X.; Yin, Y.; Zhang, J.W.; Zhang, B.B.; Bian, Z.H.; Quan, C.; Zhou, L.Y.; Hu, Y.L.; Wang, Q.F.; Ni, S.J.; et al. miR-139–5p inhibits migration and invasion of colorectal cancer by downregulating AMFR and NOTCH1. Protein Cell 2014, 5, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Wiklund, E.D.; Bramsen, J.B.; Hulf, T.; Dyrskjot, L.; Ramanathan, R.; Hansen, T.B.; Villadsen, S.B.; Gao, S.; Ostenfeld, M.S.; Borre, M.; et al. Coordinated epigenetic repression of the miR-200 family and miR-205 in invasive bladder cancer. Int. J. Cancer 2011, 128, 1327–1334. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, K.; Steptoe, A.L.; Martin, H.C.; Pattabiraman, D.R.; Nones, K.; Waddell, N.; Mariasegaram, M.; Simpson, P.T.; Lakhani, S.R.; Vlassov, A.; et al. miR-139–5p is a regulator of metastatic pathways in breast cancer. RNA 2013, 19, 1767–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchida, Y.; Chiyomaru, T.; Enokida, H.; Kawakami, K.; Tatarano, S.; Kawahara, K.; Nishiyama, K.; Seki, N.; Nakagawa, M. miR-133a induces apoptosis through direct regulation of GSTP1 in bladder cancer cell lines. Urol. Oncol. Semin. Orig. 2013, 31, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; An, H.Y.; Wang, B.; Liao, Q.; Li, W.D.; Jin, X.J.; Cui, S.Z.; Zhang, Y.J.; Ding, Y.Q.; Zhao, L. miR-133a represses tumour growth and metastasis in colorectal cancer by targeting lim and SH3 protein 1 and inhibiting the mapk pathway. Eur. J. Cancer 2013, 49, 3924–3935. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Tang, H.L.; Chen, B.; He, Z.M.; Deng, M.; Wu, M.Q.; Liu, X.P.; Yang, L.; Ye, F.; Xie, X.M. miR-26a suppresses tumour proliferation and metastasis by targeting metadherin in triple negative breast cancer. Cancer Lett. 2015, 357, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jiang, Y.; Yin, Y.; Li, Q.; He, J.; Jing, Y.; Qi, Y.T.; Xu, Q.; Li, W.; Lu, B.; et al. A regulatory circuit of miR-148a/152 and dnmt1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J. Mol. Cell Biol. 2013, 5, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Wang, Z.W.; Ali, S.; Ahmad, A.; Azmi, A.S.; Sarkar, S.H.; Banerjee, S.; Kong, D.J.; Li, Y.W.; Thakur, S.; et al. Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating mirnas in pancreatic cancer cells. Cancer Prev. Res. 2012, 5, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Yu, L.; Qin, D.D.; Huang, R.; Jiang, X.C.; Zou, C.D.; Tang, Q.C.; Chen, Y.G.; Wang, G.Y.; Wang, X.S.; et al. Role of microRNA-30c targeting ADAM19 in colorectal cancer. PLoS ONE 2015, 10, e0120698. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, S.K.; Kannisto, E.; Yendamuri, S. Overexpression of microRNA miR-30a or miR-191 in a549 lung cancer or BEAS-2B normal lung cell lines does not alter phenotype. PLoS ONE 2010, 5, e9219. [Google Scholar] [CrossRef] [PubMed]
- Li, L. Regulatory mechanisms and clinical perspectives of miR-34a in cancer. J. Cancer Res. Ther. 2014, 10, 805–810. [Google Scholar] [CrossRef] [PubMed]
- Misso, G.; di Martino, M.T.; de Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gulla, A.; Tagliaferri, P.; Tassone, P.; et al. miR-34: A new weapon against cancer? Mol. Ther. Nucleic Acids 2014, 3, e194. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.L.; Wang, X.Y.; Wen, C.Y.; Yang, X.L.; Song, M.M.; Chen, J.X.; Wang, C.L.; Zhang, B.; Wang, L.; Iwamoto, A.; et al. Hsa-miR-19a is associated with lymph metastasis and mediates the TNF-α induced epithelial-to-mesenchymal transition in colorectal cancer. Sci. Rep. 2015, 5, 13350. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, K.; Ito, S.; Hanafusa, H.; Shimizu, K.; Ouchida, M. Uncovering direct targets of miR-19a involved in lung cancer progression. PLoS ONE 2015, 10, e0137887. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.L.; Jiang, J.K.; Yang, S.H.; Huang, T.S.; Lan, H.Y.; Teng, H.W.; Yang, C.Y.; Tsai, Y.P.; Lin, C.H.; Wang, H.W.; et al. MicroRNA-146a directs the symmetric division of snail-dominant colorectal cancer stem cells. Nat. Cell Biol. 2014, 16, 383–383. [Google Scholar] [CrossRef]
- Jiang, W.; Gu, W.; Qiu, R.; Shen, C.; YaohaoWu, E.Y.; Zhang, J.; Zhou, J.; Guo, Y.; Li, Z.; Deng, J.; et al. MiRNA-101 suppresses epithelial-to-mesenchymal transition by targeting HMGA2 in pancreatic cancer cells. Anti-Cancer Agents Med. Chem. 2016, 16, 432–439. [Google Scholar] [CrossRef]
- Cornett, A.L.; Lutz, C.S. Regulation of cox-2 expression by miR-146a in lung cancer cells. RNA 2014, 20, 1419–1430. [Google Scholar] [CrossRef] [PubMed]
- Smith, A.R.; Marquez, R.T.; Tsao, W.C.; Pathak, S.; Roy, A.; Ping, J.; Wilkerson, B.; Lan, L.; Meng, W.J.; Neufeld, K.L.; et al. Tumor suppressive microRNA-137 negatively regulates musashi-1 and colorectal cancer progression. Oncotarget 2015, 6, 12558–12573. [Google Scholar] [CrossRef] [PubMed]
- Xiu, Y.C.; Liu, Z.; Xia, S.Y.; Jin, C.J.; Yin, H.F.; Zhao, W.M.; Wu, Q. MicroRNA-137 upregulation increases bladder cancer cell proliferation and invasion by targeting PAQR3. PLoS ONE 2014, 9, e109734. [Google Scholar] [CrossRef] [PubMed]
- Selth, L.A.; Townley, S.L.; Bert, A.G.; Stricker, P.D.; Sutherland, P.D.; Horvath, L.G.; Goodall, G.J.; Butler, L.M.; Tilley, W.D. Circulating micrornas predict biochemical recurrence in prostate cancer patients. Br. J. Cancer 2013, 109, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhao, C.Y.; Zhang, S.H.; Yu, D.H.; Chen, Y.; Liu, Q.H.; Shi, M.; Ni, C.R.; Zhu, M.H. Upregulation of miR-194 contributes to tumor growth and progression in pancreatic ductal adenocarcinoma. Oncol. Rep. 2014, 31, 1157–1164. [Google Scholar] [PubMed]
- Fan, Y.; Song, X.; Du, H.; Luo, C.; Wang, X.; Yang, X.; Wang, Y.; Wu, X. Down-regulation of miR-29c in human bladder cancer and the inhibition of proliferation in T24 cell via PI3K-AKT pathway. Med. Oncol. 2014, 31, 65. [Google Scholar] [CrossRef] [PubMed]
- Duhachek-Muggy, S.; Zolkiewska, A. Adam12-l is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer 2015, 15, 1. [Google Scholar] [CrossRef] [PubMed]
- Taylor, M.A.; Sossey-Alaoui, K.; Thompson, C.L.; Danielpour, D.; Schiemann, W.P. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J. Clin. Investig. 2013, 123, 150–163. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.H.; Xu, D.; Wang, Q.G.; Zheng, D.T.; Jiang, X.Q.; Xu, L.J. LPS induced miR-181a promotes pancreatic cancer cell migration via targeting PTEN and MAP2K4. Dig. Dis. Sci. 2014, 59, 1452–1460. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Shao, G.; Lv, X.; Liu, Y.; Fan, Y.; Wu, A.; Hu, H. Downregulation of miRNA-128 sensitises breast cancer cell to chemodrugs by targeting bax. Cell Biol. Int. 2013, 37, 653–658. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Zhang, T.; Liu, C.; Badeaux, M.A.; Liu, B.; Liu, R.; Jeter, C.; Chen, X.; Vlassov, A.V.; Tang, D.G. miRNA-128 suppresses prostate cancer by inhibiting BMI-1 to inhibit tumor-initiating cells. Cancer Res. 2014, 74, 4183–4195. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Yang, S.; Liu, T.; Lou, G. miR-211 suppresses epithelial ovarian cancer proliferation and cell-cycle progression by targeting cyclin d1 and cdk6. Mol. Cancer 2015, 14, 57. [Google Scholar] [CrossRef] [PubMed]
- Li, W.D.; Jin, X.J.; Zhang, Q.B.; Zhang, G.; Deng, X.B.; Ma, L. Decreased expression of miR-204 is associated with poor prognosis in patients with breast cancer. Int. J. Clin. Exp. Pathol. 2014, 7, 3287–3292. [Google Scholar] [PubMed]
- Doberstein, K.; Steinmeyer, N.; Hartmetz, A.K.; Eberhardt, W.; Mittelbronn, M.; Harter, P.N.; Juengel, E.; Blaheta, R.; Pfeilschifter, J.; Gutwein, P. MicroRNA-145 targets the metalloprotease ADAM17 and is suppressed in renal cell carcinoma patients. Neoplasia 2013, 15, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Kou, B.; Gao, Y.; Du, C.; Shi, Q.; Xu, S.; Wang, C.Q.; Wang, X.Y.; He, D.L.; Guo, P. miR-145 inhibits invasion of bladder cancer cells by targeting PAK1. Urol. Oncol. Semin. Orig. 2014, 32, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.W.; Fu, S.; Liu, Y.B.; Xu, Z.H.; Liu, Y.; Hao, J.W.; Wang, B.C.; Zhang, A.M. miR-96 suppresses renal cell carcinoma invasion via downregulation of ezrin expression. J. Exp. Clin. Cancer Res. 2015, 34, 1. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Yu, J.B.; Pan, X.L.; Li, Z.L.; Chen, Z.; Zhang, W.J.; Wang, B.; Yang, L.; Xu, H.; Zhang, G.X.; et al. HERG1 functions as an oncogene in pancreatic cancer and is downregulated by miR-96. Oncotarget 2014, 5, 5832–5844. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Yu, F.; Yao, H.; Cui, X.; Jiao, Y.; Lin, L.; Chen, J.; Yin, D.; Song, E.; Liu, Q. miR-27a regulates endothelial differentiation of breast cancer stem like cells. Oncogene 2014, 33, 2629–2638. [Google Scholar] [CrossRef] [PubMed]
- Li, J.P.; Wang, Y.P.; Song, Y.L.; Fu, Z.M.; Yu, W.J. miR-27a regulates cisplatin resistance and metastasis by targeting RKIP in human lung adenocarcinoma cells. Mol. Cancer 2014, 13, 1. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Bassi, C.; Pedriali, M.; Pagotto, S.; D’Abundo, L.; Zagatti, B.; Corra, F.; Musa, G.; Callegari, E.; Lupini, L.; et al. miR-125b targets erythropoietin and its receptor and their expression correlates with metastatic potential and ERBB2/HER2 expression. Mol. Cancer 2013, 12, 130. [Google Scholar] [CrossRef] [PubMed]
- Cortez, M.A.; Valdecanas, D.; Zhang, X.C.; Zhan, Y.A.; Bhardwaj, V.; Calin, G.A.; Komaki, R.; Giri, D.K.; Quini, C.C.; Wolfe, T.; et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol. Ther. 2014, 22, 1494–1503. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.S.; Hao, Y.; Yang, J.Y.; Zhou, Y.; Li, J.; Yin, S.Y.; Sun, C.H.; Ma, M.; Huang, Y.Y.; Xi, J.J. Genome-wide functional screening of miR-23b as a pleiotropic modulator suppressing cancer metastasis. Nat. Commun. 2011, 2, 554. [Google Scholar] [CrossRef] [PubMed]
- Li, W.P.; Liu, Z.Y.; Chen, L.; Zhou, L.; Yao, Y.Q. MicroRNA-23b is an independent prognostic marker and suppresses ovarian cancer progression by targeting runt-related transcription factor-2. FEBS Lett. 2014, 588, 1608–1615. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An analysis of human microRNA and disease associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, X.F.; Xi, Y. Normalizing bead-based microrna expression data: A measurement error model-based approach. Bioinformatics 2011, 27, 1506–1512. [Google Scholar] [CrossRef] [PubMed]




| MircoRNA | Cancer | Validation from Other References |
|---|---|---|
| miR-17, miR-20, miR-106a, miR-106b | Bladder | [20,21] |
| miR-92, miR-25 | Bladder | [22,23] |
| miR-16, miR-195 | Bladder | [24,25] |
| miR-143 | Pancreatic | [26,27] |
| miR-99a, miR-99b, miR-100 | Pancreatic, Lung | [28,29] |
| miR-103, miR-107 | Pancreatic, Lung | [30,31] |
| miR-193 | Colon, Breast | [32,33] |
| miR-205 | Pancreatic, Bladder | [34,35] |
| miR-139 | Colon, Breast, Bladder | [34,36] |
| miR-133a | Colon, Pancreatic, Bladder | [37,38] |
| miR-152, miR-148 | Lung, Breast, Bladder | [39,40] |
| miR-26a, miR-26b | Pancreatic, Breast, Bladder | [39,41] |
| miR-30a, miR-30b, miR-30c, miR-30d, miR-30e | Colon, Pancreatic, Lung, Bladder | [42,43] |
| miR-34b, miR-34c | Colon, Lung, Breast, Kidney | [44,45] |
| miR-19a, miR-19b | Colon, Lung, Breast, Bladder | [46,47] |
| miR-101 | Colon, Pancreatic, Breast, Kidney | [48,49] |
| miR-146 | Colon, Lung, Bladder, Kidney | [48,50] |
| miR-137 | Colon, Pancreatic, Bladder, Kidney | [51,52] |
| miR-194 | Prostate, Pancreatic, Lung, Bladder | [53,54] |
| miR-29a, miR-29b, miR-29c | Prostate, Breast, Bladder, Kidney | [55,56] |
| miR-181a, miR-181c | Pancreatic, Breast, Bladder, Kidney | [57,58] |
| miR-128a, miR-128b | Prostate, Pancreatic, Breast, Bladder | [59,60] |
| miR-204, miR-211 | Pancreatic, Breast, Bladder, Kidney | [61,62] |
| miR-145 | Colon, Pancreatic, Breast, Bladder, Kidney | [63,64] |
| miR-96 | Colon, Prostate, Pancreatic, Breast, Kidney | [65,66] |
| miR-27a, miR-27b | Prostate, Pancreatic, Lung, Breast, Bladder, Kidney | [67,68] |
| miR-125a, miR-125b, miR-200b | Colon, Prostate, Pancreatic, Lung, Breast, Bladder, Kidney | [69,70] |
| miR-23a, miR-23b | Colon, Prostate, Pancreatic, Lung, Breast, Bladder, Kidney | [71,72] |
| miRNAs Selected in [9] | miRNAs Confirmed by HMDD | miRNAs Selected by the Method | miRNAs Selected by the Method and Confirmed by HMDD | Sensitivity | Specificity | |
|---|---|---|---|---|---|---|
| A | B | C | D | D/B | (C − D)/(A − B) | |
| Colon | 42 | 21 | 19 | 10 | 0.476 | 0.429 |
| Pancreatic | 54 | 29 | 33 | 17 | 0.586 | 0.64 |
| Prostate | 24 | 2 | 14 | 1 | 0.5 | 0.591 |
| Lung | 41 | 34 | 25 | 20 | 0.588 | 0.714 |
| Breast | 49 | 42 | 29 | 26 | 0.619 | 0.429 |
| Bladder | 70 | 34 | 42 | 17 | 0.5 | 0.694 |
| Kidney | 37 | 10 | 21 | 6 | 0.6 | 0.556 |
| Colon | Prostate | Pancreatic | Lung | Breast | Bladder | Kidney | Group Number in Figure 2 | |
|---|---|---|---|---|---|---|---|---|
| miR-124a | v | v | v | v | v | v | 1 | |
| miR-9 | v | v | v | v | v | v | 1 | |
| miR-182 | v | v | v | v | v | v | 1 | |
| miR-135 | v | v | v | v | v | v | 1 | |
| miR-125b | v | v | v | v | v | v | v | 2 |
| miR-23b | v | v | v | v | v | v | v | 2 |
| miR-23a | v | v | v | v | v | v | v | 2 |
| miR-125a | v | v | v | v | v | v | v | 2 |
| miR-200b | v | v | v | v | v | v | v | 2 |
| miR-200c | v | v | v | v | v | v | v | 2 |
| miR-146 | v | v | v | v | 3 | |||
| miR-199 | v | v | v | v | v | 4 | ||
| miR-1 | v | v | 5 | |||||
| miR-30b | v | v | v | v | 6 | |||
| miR-200a | v | v | v | v | 6 | |||
| miR-30a | v | v | v | v | 6 | |||
| miR-30d | v | v | v | v | 6 | |||
| miR-30c | v | v | v | v | 6 | |||
| miR-30e | v | v | v | v | 6 | |||
| miR-137 | v | v | v | v | 7 | |||
| miR-19a | v | v | v | v | 8 | |||
| miR-19b | v | v | v | v | 8 | |||
| miR-203 | v | v | v | v | v | 9 | ||
| miR-155 | v | v | v | v | v | 9 | ||
| miR-33 | v | v | v | v | v | 10 | ||
| miR-219 | v | 11 | ||||||
| miR-216 | v | 11 | ||||||
| miR-223 | v | v | v | v | 12 | |||
| miR-193 | v | v | 13 | |||||
| miR-29b | v | v | v | v | 14 | |||
| miR-29c | v | v | v | v | 14 | |||
| miR-29a | v | v | v | v | 14 | |||
| miR-206 | v | v | v | 15 | ||||
| miR-218 | v | v | v | v | 16 | |||
| miR-128b | v | v | v | v | 16 | |||
| miR-128a | v | v | v | v | 16 | |||
| miR-34a | v | v | v | v | 17 | |||
| miR-34b | v | v | v | v | 17 | |||
| miR-34c | v | v | v | v | 17 | |||
| miR-194 | v | v | v | v | 18 | |||
| miR-138 | v | v | v | 19 | ||||
| miR-96 | v | v | v | v | v | 20 | ||
| miR-27b | v | v | v | v | v | v | 21 | |
| miR-27a | v | v | v | v | v | v | 21 | |
| miR-99a | v | v | 22 | |||||
| miR-100 | v | v | 22 | |||||
| miR-107 | v | v | 22 | |||||
| miR-103 | v | v | 22 | |||||
| miR-99b | v | v | 22 | |||||
| miR-181a | v | v | v | v | 23 | |||
| miR-181b | v | v | v | v | 23 | |||
| miR-204 | v | v | v | v | 23 | |||
| miR-211 | v | v | v | v | 23 | |||
| miR-181c | v | v | v | v | 23 | |||
| miR-24 | v | v | v | v | v | v | 24 | |
| miR-205 | v | v | 25 | |||||
| miR-215 | v | v | v | 26 | ||||
| miR-192 | v | v | v | 26 | ||||
| miR-21 | v | v | v | v | v | 27 | ||
| miR-190 | v | v | v | 28 | ||||
| miR-26a | v | v | v | 28 | ||||
| miR-144 | v | v | v | 28 | ||||
| miR-26b | v | v | v | 28 | ||||
| miR-183 | v | v | v | v | v | 29 | ||
| miR-22 | v | v | v | v | v | 30 | ||
| miR-145 | v | v | v | v | v | 31 | ||
| miR-140 | v | v | 32 | |||||
| miR-139 | v | v | v | 33 | ||||
| miR-143 | v | 34 | ||||||
| miR-133a | v | v | v | 35 | ||||
| miR-18 | v | 36 | ||||||
| miR-101 | v | v | v | v | 37 | |||
| miR-152 | v | v | v | 38 | ||||
| miR-148 | v | v | v | 38 | ||||
| miR-141 | v | v | v | 38 | ||||
| miR-302 | v | v | 39 | |||||
| miR-93 | v | v | 39 | |||||
| miR-16 | v | 40 | ||||||
| miR-92 | v | 40 | ||||||
| miR-142 | v | 40 | ||||||
| miR106a | v | 40 | ||||||
| miR-17 | v | 40 | ||||||
| miR-195 | v | 40 | ||||||
| miR-20 | v | 40 | ||||||
| miR-15a | v | 40 | ||||||
| miR-153 | v | 40 | ||||||
| miR-106b | v | 40 | ||||||
| miR-25 | v | 40 | ||||||
| miR-32 | v | 40 | ||||||
| miR-199b | v | 41 |
© 2016 by the author; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H. Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis. Int. J. Mol. Sci. 2016, 17, 773. https://doi.org/10.3390/ijms17050773
Wang H. Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis. International Journal of Molecular Sciences. 2016; 17(5):773. https://doi.org/10.3390/ijms17050773
Chicago/Turabian StyleWang, Hsiuying. 2016. "Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis" International Journal of Molecular Sciences 17, no. 5: 773. https://doi.org/10.3390/ijms17050773
APA StyleWang, H. (2016). Predicting MicroRNA Biomarkers for Cancer Using Phylogenetic Tree and Microarray Analysis. International Journal of Molecular Sciences, 17(5), 773. https://doi.org/10.3390/ijms17050773