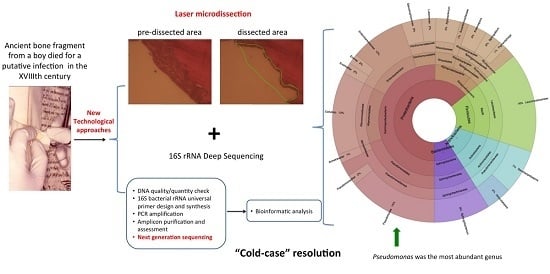
The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing
Abstract
:1. Introduction
2. Results
3. Discussion
4. Materials and Methods
4.1. Sample Obtainment and Preparation for Analysis
4.2. Hematoxylin and Eosin Staining
4.3. Laser Microdissection and DNA Extraction
4.4. Sex-Determining Region Y Amplification
4.5. Microbiome Typing by 16S rRNA Deep Sequencing
4.6. Bioinformatics
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| H&E | Hematoxylin and eosin |
| SRY | Sex-determining region Y |
| OTUs | Operational taxonomic units |
References
- Lugli, A.; Clemenza, M.; Corso, P.E.; di Costanzo, J.; Dirnhofer, R.; Fiorini, E.; Herborg, C.; Hindmarsh, J.T.; Orvini, E.; Piazzoli, A.; et al. The medical mystery of Napoleon Bonaparte: An interdisciplinary exposé. Adv. Anat. Pathol. 2011, 18, 152–158. [Google Scholar] [CrossRef] [PubMed]
- Gallassi, F.M.; Ashrafian, H. Has the diagnosis of a stroke been overlooked in the symptoms of Julius Caesar? Neurol. Sci. 2015, 36, 1521–1522. [Google Scholar] [CrossRef] [PubMed]
- Zink, A.R.; Reischl, U.; Wolf, H.; Nerlich, A.G. Molecular analysis of ancient microbial infections. FEMS Microbiol. Lett. 2002, 213, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Padmanabhan, R.; Mishra, A.K.; Raoult, D.; Fournier, P.E. Genomics and metagenomics in medical microbiology. J. Microbiol. Methods 2013, 95, 415–424. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Salvatore, F. The role of the gut microbiome in the healthy adult status. Clin. Chim. Acta 2015, 451, 97–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, K. Pseudomonas aeruginosa: Evolution of antimicrobial resistance and implications for therapy. Semin. Respir. Crit. Care Med. 2015, 36, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Lenney, W. Pseudomonas aeruginosa in cystic fibrosis is potentially serious, and more than merely a marker for disease severity. Paediatr. Respir. Rev. 2015, 16, 35–36. [Google Scholar] [CrossRef] [PubMed]
- Aydın, N.; Şirin, E.; Aydemir, A.N.; Zengin, G. Pseudomonas osteomyelitis of the proximal humerus after arthroscopic rotator cuff repair. Acta Orthop. Traumatol. Turc. 2014, 48, 685–689. [Google Scholar] [CrossRef] [PubMed]
- Anzai, Y.; Kim, H.; Park, J.Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50, 1563–1589. [Google Scholar] [CrossRef] [PubMed]
- Barahona, F.; Slim, J. Sphingobacterium multivorum: Case report and literature review. New Microbes New Infect. 2015, 7, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Gardner, J.G. Polysaccharide degradation systems of the saprophytic bacterium Cellvibrio japonicus. World J. Microbiol. Biotechnol. 2016, 32, 121. [Google Scholar] [CrossRef] [PubMed]
- Van Asten, S.A.; La Fontaine, J.; Peters, E.J.; Bhavan, K.; Kim, P.J.; Lavery, L.A. The microbiome of diabetic foot osteomyelitis. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Pasyk, K.A.; Hassett, C.A. Modified hematoxylin and eosin staining method for epoxy-embedded tissue sections. Pathol. Res. Pract. 1989, 184, 635–638. [Google Scholar] [CrossRef]
- Shekhtman, E.M.; Anne, K.; Melkonyan, H.S.; Robbins, D.J.; Warsof, S.L.; Umansky, S.R. Optimization of transrenal DNA analysis: Detection of fetal DNA in maternal urine. Clin. Chem. 2009, 55, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Pagliuca, C.; Colicchio, R.; Sarnataro, D.; Discepolo, V.; Kim, S.M.; Russo, I.; del Vecchio Blanco, G.; et al. Metagenomics Reveals Dysbiosis and a Potentially Pathogenic N. flavescens Strain in Duodenum of Adult Celiac Patients. Am. J. Gastroenterol. 2016, 111, 879–890. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- D’Argenio, V.; Casaburi, G.; Precone, V.; Salvatore, F. Comparative metagenomic analysis of human gut microbiome composition using two different bioinformatic pipelines. BioMed Res. Int. 2014, 2014, 325340. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- De Santis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]



| Phylum | Class | Order | Family | Genus | Relative Abundance at Genus Level (%) | Number of High Quality Reads at Genus Level |
|---|---|---|---|---|---|---|
| Actinobacteria | Actinobacteria | Actinomycetales | Brevibacteriaceae | Brevibacterium | 0.31 | 9 |
| Actinobacteria | Actinobacteria | Actinomycetales | Corynebacteriaceae | Corynebacterium | 0.75 | 22 |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Agrococcus | 0.38 | 11 |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Leucobacter | 0.10 | 3 |
| Actinobacteria | Actinobacteria | Actinomycetales | Microbacteriaceae | Microbacterium | 0.68 | 20 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | Arthrobacter | 0.21 | 6 |
| Actinobacteria | Actinobacteria | Actinomycetales | Micrococcaceae | Micrococcus | 0.96 | 28 |
| Actinobacteria | Actinobacteria | Actinomycetales | Propionibacteriaceae | Propionibacterium | 1.34 | 39 |
| Actinobacteria | Actinobacteria | Actinomycetales | Pseudonocardiaceae | Amycolatopsis | 5.14 | 150 |
| Actinobacteria | Actinobacteria | Actinomycetales | Pseudonocardiaceae | Pseudonocardia | 1.13 | 33 |
| Actinobacteria | Actinobacteria | Actinomycetales | Pseudonocardiaceae | Saccharopolyspora | 8.87 | 259 |
| Bacteroidetes | Flavobacteriia | Flavobacteriales | Flavobacteriaceae | Flavobacterium | 0.41 | 12 |
| Bacteroidetes | Sphingobacteriia | Sphingobacteriales | Sphingobacteriaceae | Sphingobacterium | 11.26 | 329 |
| Firmicutes | Bacilli | Bacillales | Bacillaceae | Bacillus | 0.72 | 21 |
| Firmicutes | Bacilli | Bacillales | Planococcaceae | Lysinibacillus | 0.14 | 4 |
| Firmicutes | Bacilli | Bacillales | Planococcaceae | Rummeliibacillus | 0.79 | 23 |
| Firmicutes | Bacilli | Bacillales | Staphylococcaceae | Staphylococcus | 0.45 | 13 |
| Firmicutes | Bacilli | Lactobacillales | Aerococcaceae | Aerococcus | 0.79 | 23 |
| Firmicutes | Bacilli | Lactobacillales | Enterococcaceae | Enterococcus | 0.07 | 2 |
| Firmicutes | Bacilli | Lactobacillales | Streptococcaceae | Streptococcus | 0.45 | 13 |
| Proteobacteria | Alphaproteobacteria | Caulobacterales | Caulobacteraceae | Brevundimonas | 0.10 | 3 |
| Proteobacteria | Alphaproteobacteria | Rhizobiales | Rhizobiaceae | Agrobacterium | 4.86 | 142 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Anaerospora | 3.87 | 113 |
| Proteobacteria | Alphaproteobacteria | Rhodobacterales | Rhodobacteraceae | Rhodobacter | 1.10 | 32 |
| Proteobacteria | Alphaproteobacteria | Sphingomonadales | Sphingomonadaceae | Sphingobium | 3.22 | 94 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Achromobacter | 0.27 | 8 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Alcaligenaceae | Pigmentiphaga | 1.64 | 48 |
| Proteobacteria | Betaproteobacteria | Burkholderiales | Oxalobacteraceae | Janthinobacterium | 0.10 | 3 |
| Proteobacteria | Gammaproteobacteria | Alteromonadales | Alteromonadaceae | Cellvibrio | 10.50 | 482 |
| Proteobacteria | Gammaproteobacteria | Enterobacteriales | Enterobacteriaceae | Erwinia | 4.59 | 134 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Moraxellaceae | Acinetobacter | 1.92 | 56 |
| Proteobacteria | Gammaproteobacteria | Pseudomonadales | Pseudomonadaceae | Pseudomonas | 15.00 | 584 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Luteimonas | 2.94 | 86 |
| Proteobacteria | Gammaproteobacteria | Xanthomonadales | Xanthomonadaceae | Stenotrophomonas | 3.97 | 116 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Argenio, V.; Torino, M.; Precone, V.; Casaburi, G.; Esposito, M.V.; Iaffaldano, L.; Malapelle, U.; Troncone, G.; Coto, I.; Cavalcanti, P.; et al. The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing. Int. J. Mol. Sci. 2017, 18, 109. https://doi.org/10.3390/ijms18010109
D’Argenio V, Torino M, Precone V, Casaburi G, Esposito MV, Iaffaldano L, Malapelle U, Troncone G, Coto I, Cavalcanti P, et al. The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing. International Journal of Molecular Sciences. 2017; 18(1):109. https://doi.org/10.3390/ijms18010109
Chicago/Turabian StyleD’Argenio, Valeria, Marielva Torino, Vincenza Precone, Giorgio Casaburi, Maria Valeria Esposito, Laura Iaffaldano, Umberto Malapelle, Giancarlo Troncone, Iolanda Coto, Paolina Cavalcanti, and et al. 2017. "The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing" International Journal of Molecular Sciences 18, no. 1: 109. https://doi.org/10.3390/ijms18010109
APA StyleD’Argenio, V., Torino, M., Precone, V., Casaburi, G., Esposito, M. V., Iaffaldano, L., Malapelle, U., Troncone, G., Coto, I., Cavalcanti, P., De Rosa, G., Salvatore, F., & Sacchetti, L. (2017). The Cause of Death of a Child in the 18th Century Solved by Bone Microbiome Typing Using Laser Microdissection and Next Generation Sequencing. International Journal of Molecular Sciences, 18(1), 109. https://doi.org/10.3390/ijms18010109