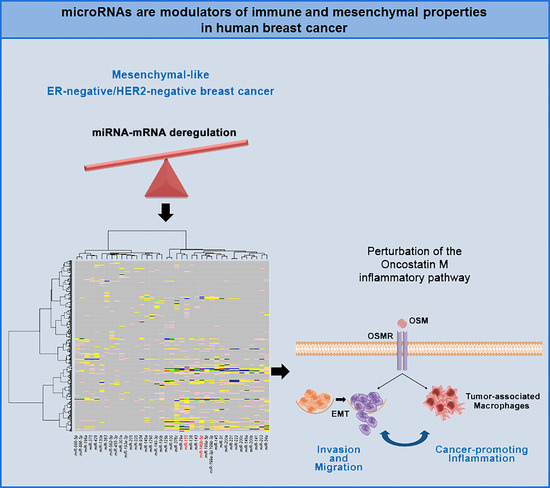
Integrated MicroRNA–mRNA Profiling Identifies Oncostatin M as a Marker of Mesenchymal-Like ER-Negative/HER2-Negative Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. Specific miRNA Expression Patterns Define Molecularly Different Human Breast Cancer Cell Lines
2.2. Correlation between miRNA and mRNA Expression Profiles and Pathway Analysis
2.3. Oncostatin M Expression is Associated with ER-Negative/HER2-Negative Breast Cancer
3. Discussion
4. Materials and Methods
4.1. Cell Culture, RNA Isolation, and Microarray Experiments
4.2. Human Breast Cancer Datasets and in Silico Analysis
4.3. Tumor Samples and Immunohistochemistry
4.4. Statistics and Bioinformatics
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interests
References
- Banerji, S.; Cibulskis, K.; Rangel-Escareno, C.; Brown, K.K.; Carter, S.L.; Frederick, A.M.; Lawrence, M.S.; Sivachenko, A.Y.; Sougnez, C.; Zou, L.; et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 2012, 486, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, L.; Qi, Y.; Stemke-Hale, K.; Wang, B.; Young, E.J.; Booser, D.J.; Holmes, F.A.; O’Shaughnessy, J.; Hellerstedt, B.; Pippen, J.; et al. Mutation profiling identifies numerous rare drug targets and distinct mutation patterns in different clinical subtypes of breast cancers. Breast Cancer Res. Treat. 2012, 134, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Iwamoto, T.; Bianchini, G.; Booser, D.; Qi, Y.; Coutant, C.; Shiang, C.Y.; Santarpia, L.; Matsuoka, J.; Hortobagyi, G.N.; Symmans, W.F.; et al. Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J. Natl. Cancer Inst. 2011, 103, 264–272. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B.; Bottai, G.; Fleischer, T.; Munkácsy, G.; Budczies, J.; Paladini, L.; Børresen-Dale, A.L.; Kristensen, V.N.; Santarpia, L. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int. J. Cancer 2016, 138, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Santarpia, L.; Iwamoto, T.; di Leo, A.; Hayashi, N.; Bottai, G.; Stampfer, M.; Andre, F.; Turner, N.C.; Symmans, W.F.; Hortobágyi, G.N.; et al. DNA repair gene patterns as prognostic and predictive factors in molecular breast cancer subtypes. Oncologist 2013, 18, 1063–1073. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.; Shah, S.P.; Chin, S.F.; Turashvili, G.; Rueda, O.M.; Dunning, M.J.; Speed, D.; Lynch, A.G.; Samarajiwa, S.; Yuan, Y.; et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature 2012, 486, 346–352. [Google Scholar] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef] [PubMed]
- Turner, N.C.; Reis-Filho, J.S. Tackling the diversity of triple-negative breast cancer. Clin. Cancer Res. 2013, 19, 6380–6388. [Google Scholar] [CrossRef] [PubMed]
- Fridman, W.H.; Pagès, F.; Sautès-Fridman, C.; Galon, J. The immune contexture in human tumours: Impact on clinical outcome. Nat. Rev. Cancer 2012, 12, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Savas, P.; Salgado, R.; Denkert, C.; Sotiriou, C.; Darcy, P.K.; Smyth, M.J.; Loi, S. Clinical relevance of host immunity in breast cancer: From TILs to the clinic. Nat. Rev. Clin. Oncol. 2016, 13, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Croce, C.M. MicroRNA dysregulation in cancer: Diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol. Med. 2012, 4, 143–159. [Google Scholar] [CrossRef] [PubMed]
- Hausser, J.; Zavolan, M. Identification and consequences of miRNA-target interactions—Beyond repression of gene expression. Nat. Rev. Genet. 2014, 15, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Kohlhapp, F.J.; Mitra, A.K.; Lengyel, E.; Peter, M.E. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 2015, 34, 5857–5868. [Google Scholar] [CrossRef] [PubMed]
- Paladini, L.; Fabris, L.; Bottai, G.; Raschioni, C.; Calin, G.A.; Santarpia, L. Targeting microRNAs as key modulators of tumor immune response. J. Exp. Clin. Cancer Res. 2016, 35, 103. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Blenkiron, C.; Goldstein, L.D.; Thorne, N.P.; Spiteri, I.; Chin, S.F.; Dunning, M.J.; Barbosa-Morais, N.L.; Teschendorff, A.E.; Green, A.R.; Ellis, I.O.; et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007, 8, R214. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microRNA expression profiling identifies miR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Galasso, M.; Sana, M.E.; Wise, T.F.; Palatini, J.; Huebner, K.; Croce, C.M. Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. USA 2012, 109, 3024–3029. [Google Scholar] [CrossRef] [PubMed]
- Michaut, M.; Chin, S.F.; Majewski, I.; Severson, T.M.; Bismeijer, T.; de Koning, L.; Peeters, J.K.; Schouten, P.C.; Rueda, O.M.; Bosma, A.J.; et al. Integration of genomic, transcriptomic and proteomic data identifies two biologically distinct subtypes of invasive lobular breast cancer. Sci. Rep. 2016, 6, 18517. [Google Scholar] [CrossRef] [PubMed]
- Hamed, M.; Spaniol, C.; Zapp, A.; Helms, V. Integrative network-based approach identifies key genetic elements in breast invasive carcinoma. BMC Genom. 2015, 16, S2. [Google Scholar] [CrossRef] [PubMed]
- De Rinaldis, E.; Gazinska, P.; Mera, A.; Modrusan, Z.; Fedorowicz, G.M.; Burford, B.; Gillett, C.; Marra, P.; Grigoriadis, A.; Dornan, D.; et al. Integrated genomic analysis of triple-negative breast cancers reveals novel microRNAs associated with clinical and molecular phenotypes and sheds light on the pathways they control. BMC Genom. 2013, 14, 643. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, V.N.; Vaske, C.J.; Ursini-Siegel, J.; van Loo, P.; Nordgard, S.H.; Sachidanandam, R.; Sørlie, T.; Wärnberg, F.; Haakensen, V.D.; Helland, Å.; et al. Integrated molecular profiles of invasive breast tumors and ductal carcinoma in situ (DCIS) reveal differential vascular and interleukin signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 2802–2807. [Google Scholar] [CrossRef] [PubMed]
- Volinia, S.; Croce, C.M. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 7413–7417. [Google Scholar] [CrossRef] [PubMed]
- Buffa, F.M.; Camps, C.; Winchester, L.; Snell, C.E.; Gee, H.E.; Sheldon, H.; Taylor, M.; Harris, A.L.; Ragoussis, J. MicroRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011, 71, 5635–5645. [Google Scholar] [CrossRef] [PubMed]
- Luo, D.; Wilson, J.M.; Harvel, N.; Liu, J.; Pei, L.; Huang, S.; Hawthorn, L.; Shi, H. A systematic evaluation of miRNA:mRNA interactions involved in the migration and invasion of breast cancer cells. J. Transl. Med. 2013, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Chen, A.; Bai, Z. Integrative investigation on breast cancer in ER, PR and HER2-defined subgroups using mRNA and miRNA expression profiling. Sci. Rep. 2014, 4, 6566. [Google Scholar] [CrossRef] [PubMed]
- Joshi, T.; Elias, D.; Stenvang, J.; Alves, C.L.; Teng, F.; Lyng, M.B.; Lykkesfeldt, A.E.; Brünner, N.; Wang, J.; Gupta, R.; et al. Integrative analysis of miRNA and gene expression reveals regulatory networks in tamoxifen-resistant breast cancer. Oncotarget 2016, 7, 57239–57253. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Murphy, L.C.; Watson, P.H. Oncostatin M suppresses oestrogen receptor-α expression and is associated with poor outcome in human breast cancer. Endocr. Relat. Cancer 2012, 19, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Douglas, A.M.; Grant, S.L.; Goss, G.A.; Clouston, D.R.; Sutherland, R.L.; Begley, C.G. Oncostatin M induces the differentiation of breast cancer cells. Int. J. Cancer 1998, 75, 64–73. [Google Scholar] [CrossRef]
- Jorcyk, C.L.; Holzer, R.G.; Ryan, R.E. Oncostatin M induces cell detachment and enhances the metastatic capacity of T-47D human breast carcinoma cells. Cytokine 2006, 33, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Chen, C.; Shi, M.; Wang, F.; Chen, X.; Diao, D.; Hu, M.; Yu, M.; Qian, L.; Guo, N. Stat3-coordinated Lin-28-let-7-HMGA2 and miR-200-ZEB1 circuits initiate and maintain oncostatin M-driven epithelial-mesenchymal transition. Oncogene 2013, 32, 5272–5282. [Google Scholar] [CrossRef] [PubMed]
- West, N.R.; Murray, J.I.; Watson, P.H. Oncostatin M promotes phenotypic changes associated with mesenchymal and stem cell-like differentiation in breast cancer. Oncogene 2014, 33, 1485–1494. [Google Scholar] [CrossRef] [PubMed]
- Vlaicu, P.; Mertins, P.; Mayr, T.; Widschwendter, P.; Ataseven, B.; Högel, B.; Eiermann, W.; Knyazev, P.; Ullrich, A. Monocytes/macrophages support mammary tumor invasivity by co-secreting lineage-specific EGFR ligands and a STAT3 activator. BMC Cancer 2013, 13, 197. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, C.; Tewari, B.N.; Kanchan, R.K.; Baghel, K.S.; Nautiyal, N.; Shrivastava, R.; Kaur, H.; Bhatt, M.L.; Bhadauria, S. Macrophages are recruited to hypoxic tumor areas and acquire a pro-angiogenic M2-polarized phenotype via hypoxic cancer cell derived cytokines Oncostatin M and Eotaxin. Oncotarget 2014, 5, 5350–5368. [Google Scholar] [CrossRef] [PubMed]
- Bottai, G.; Raschioni, C.; Székely, B.; di Tommaso, L.; Szász, A.M.; Losurdo, A.; Győrffy, B.; Ács, B.; Torrisi, R.; Karachaliou, N.; et al. AXL-associated tumor inflammation as a poor prognostic signature in chemotherapy-treated triple-negative breast cancer patients. NPJ Breast Cancer 2016, 2, 16033. [Google Scholar] [CrossRef]
- Queen, M.M.; Ryan, R.E.; Holzer, R.G.; Keller-Peck, C.R.; Jorcyk, C.L. Breast cancer cells stimulate neutrophils to produce oncostatin M: Potential implications for tumor progression. Cancer Res. 2005, 65, 8896–8904. [Google Scholar] [CrossRef] [PubMed]
- Hollmén, M.; Roudnicky, F.; Karaman, S.; Detmar, M. Characterization of macrophage—Cancer cell crosstalk in estrogen receptor positive and triple-negative breast cancer. Sci. Rep. 2015, 5, 9188. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Che, Q.; Liao, Y.; Wang, H.; Wang, J.; Chen, Z.; Wang, F.; Dai, C.; Wan, X. Oncostatin M activates STAT3 to promote endometrial cancer invasion and angiogenesis. Oncol. Rep. 2015, 34, 129–138. [Google Scholar] [CrossRef] [PubMed]




| Up-Regulated | Down-Regulated | ||
|---|---|---|---|
| miRNA | p-Value 1 | miRNA | p-Value 1 |
| miR-29a | 1.52 × 10−2 | miR-34a | 5.43 × 10−4 |
| miR-31 | 6.26 × 10−3 | miR-141 | 3.08 × 10−8 |
| miR-100 | 8.54 × 10−3 | miR-148a | 5.49 × 10−4 |
| miR-125b | 1.98 × 10−2 | miR-190b | 8.54 × 10−3 |
| miR-130a | 1.90 × 10−2 | miR-193a-3p | 6.26 × 10−3 |
| miR-138 | 4.41 × 10−3 | miR-196a | 3.08 × 10−2 |
| miR-140-3p | 1.06 × 10−2 | miR-200a | 6.32 × 10−7 |
| miR-143 | 1.06 × 10−2 | miR-200b | 3.08 × 10−8 |
| miR-145 | 2.69 × 10−2 | miR-200c | 2.85 × 10−7 |
| miR-146a | 4.01 × 10−3 | miR-203 | 1.81 × 10−2 |
| miR-146b-5p | 6.24 × 10−3 | miR-205 | 2.69 × 10−2 |
| miR-155 | 3.86 × 10−3 | miR-301a | 7.13 × 10−3 |
| miR-199a-3p/199b-3p | 9.11 × 10−3 | miR-335 | 1.05 × 10−2 |
| miR-199a-5p | 2.18 × 10−3 | miR-363 | 8.86 × 10−3 |
| miR-221 | 1.88 × 10−3 | miR-375 | 1.49 × 10−2 |
| miR-222 | 2.11 × 10−3 | miR-429 | 1.06 × 10−2 |
| miR-376c | 3.66 × 10−5 | miR-934 | 3.08 × 10−2 |
| miR-455-3p | 1.81 × 10−2 | ||
| miR-582-5p | 3.64 × 10−3 | ||
| miR-886-3p | 6.24 × 10−3 | ||
| miR-886-5p | 6.26 × 10−3 | ||
| miR-1290 | 4.41 × 10−3 | ||
| Signaling Pathway | miRNA | FDR |
|---|---|---|
| OSM signaling | miR-146b-5p, miR-155 | ≤1.00 × 10−4 |
| miR-31 | ≤5.00 × 10−4 | |
| miR-29a, miR-199a-3p/199b-3p, miR-199a-5p, miR-200a, miR-221, miR-222 | ≤5.00 × 10−3 | |
| miR-100, miR-125b, miR-143, miR-376c | ≤1.00 × 10−2 | |
| miR-34a, miR-138, miR-141, miR-145, miR-146a, miR-148a, miR-200b, miR-200c, miR-203 | ≤5.00 × 10−2 | |
| ERK/MAPK signaling | miR-138 | ≤5.00 × 10−4 |
| miR-376 | ≤1.00 × 10−3 | |
| miR-100, miR-125b, miR-155 | ≤5.00 × 10−3 | |
| miR-29a, miR-31, miR-141 | ≤1.00 × 10−2 | |
| miR-34a, miR-146b-5p, miR-200a, miR-221, miR-222 | ≤5.00 × 10−2 | |
| JAK/STAT pathway | miR-155 | ≤1.00 × 10−2 |
| Integrin signaling | miR-31, miR-138, miR-143, miR-145, miR-148a | ≤5.00 × 10−3 |
| miR-29a, miR-200c | ≤1.00 × 10−2 | |
| Interleukin-3 signaling | miR-155 | ≤5.00 × 10−4 |
| miR-31 | ≤1.00 × 10−2 | |
| miR-29a, miR-145, miR-200a, miR-376 | ≤5.00 × 10−2 | |
| Interleukin -4 signaling | miR-31, miR-128a, miR-141, miR-148a, miR-155, miR-203 | ≤5.00 × 10−2 |
| Interleukin -6 signaling | miR-140, miR-190b, miR-221, miR-222 | ≤5.00 × 10−2 |
| IFN-γ signaling | miR-146b, miR-155 | ≤5.00 × 10−3 |
| miR-31 | ≤1.00 × 10−2 | |
| EGF signaling | miR-31, miR-145, miR-155 | <5.00 × 10−2 |
| T helper differentiation pathway | miR-220a | ≤1.00 × 10−2 |
| miR-31, miR-130a, miR-145, miR-199a-5p, miR-203, miR-221, miR-375 | ≤5.00 × 10−2 | |
| Semaphorins signaling | miR-196a, miR-375, miR-429, miR-934 | ≤5.00 × 10−3 |
| miR-145, miR-199a-5p, miR-200a, miR-203 | ≤1.00 × 10−2 | |
| Leucocyte extravasation | miR-200c | ≤5.00 × 10−3 |
| miR-200b, miR-203 | ≤1.00 × 10−2 |
| Category | Gene | Spearman Coefficient | 95% CI | p-Value |
|---|---|---|---|---|
| Macrophage function and immune response | ALOX15 | 0.442 | 0.34–0.54 | <1.00 × 10−4 |
| ARG1 | 0.512 | 0.34–0.54 | <1.00 × 10−4 | |
| CCL17 | 0.218 | 0.10–0.33 | 4.00 × 10−4 | |
| CCL24 | 0.296 | 0.18–0.41 | <1.00 × 10−4 | |
| CD40LG | 0.335 | 0.22–0.44 | <1.00 × 10−4 | |
| CERK | 0.196 | 0.07–0.31 | 1.50 × 10−3 | |
| CHN2 | 0.282 | 0.16–0.39 | <1.00 × 10−4 | |
| CSF2 | 0.382 | 0.27–0.48 | <1.00 × 10−4 | |
| CSF3 | 0.451 | 0.35–0.55 | <1.00 × 10−4 | |
| CSF3R | 0.460 | 0.36–0.55 | <1.00 × 10−4 | |
| CXCL9 | −0.199 | −0.32–−0.08 | 1.20 × 10−3 | |
| CXCL10 | −0.196 | −0.31–−0.07 | 1.40 × 10−3 | |
| CXCL11 | −0.219 | −0.33–−0.01 | 3.00 × 10−4 | |
| GAS7 | 0.306 | 0.19–0.42 | <1.00 × 10−4 | |
| GBP1 | −0.192 | −0.31–−0.07 | 1.80 × 10−3 | |
| HRH1 | 0.250 | 0.13–0.36 | <1.00 × 10−4 | |
| IL1RN | 0.406 | 0.30–0.51 | <1.00 × 10−4 | |
| IL4 | 0.378 | 0.27–0.48 | <1.00 × 10−4 | |
| IL6 | 0.271 | 0.15–0.38 | <1.00 × 10−4 | |
| IL6R | 0.232 | 0.11–0.5 | 2.00 × 10−4 | |
| IL10 | 0.286 | 0.17–0.40 | <1.00 × 10−4 | |
| IL15RA | −0.228 | −0.34–−0.11 | 2.00 × 10−4 | |
| IL17A | 0.280 | 0.16–0.39 | <1.00 × 10−4 | |
| IL32 | −0.162 | −0.28–−0.04 | 8.80 × 10−3 | |
| MERTK | 0.162 | 0.04–0.28 | 8.60 × 10−3 | |
| NFKB1 | 0.385 | 0.27–0.49 | <1.00 × 10−4 | |
| TNF | 0.376 | 0.26–0.48 | <1.00 × 10−4 | |
| EMT, EGF signaling and downstream pathways | CDH1 | −0.317 | −0.42–−0.20 | <1.00 × 10−4 |
| COL1A2 | −0.338 | −0.44–−0.22 | <1.00 × 10−4 | |
| COL3A1 | −0.310 | −0.42–−0.19 | <1.00 × 10−4 | |
| DSP | −0.227 | −0.34–−0.10 | 2.00 × 10−4 | |
| EGFR | 0.371 | 0.26–0.47 | <1.00 × 10−4 | |
| MAP2K6 | 0.238 | 0.12–035 | 1.00 × 10−4 | |
| MAP2K7 | 0.466 | 0.36–056 | <1.00 × 10−4 | |
| MAP3K2 | 0.358 | 0.24–0.46 | <1.00 × 10−4 | |
| MAPK8 | 0.33 | 0.22–0.44 | <1.00 × 10−4 | |
| MAPK10 | 0.272 | 0.15–0.38 | <1.00 × 10−4 | |
| MTOR | 0.385 | 0.27–0.49 | <1.00 × 10−4 | |
| PIK3R2 | 0.213 | 0.09–0.33 | 5.00 × 10−4 | |
| RHOA | 0.250 | 0.13–0.36 | <1.00 × 10−4 | |
| SRC | 0.237 | 0.12–0.35 | 1.00 × 10−4 | |
| TGFB1 | 0.446 | 0.34–0.54 | <1.00 × 10−4 | |
| ZEB2 | 0.160 | 0.04–0.28 | 9.60 × 10−3 |
© 2017 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bottai, G.; Diao, L.; Baggerly, K.A.; Paladini, L.; Győrffy, B.; Raschioni, C.; Pusztai, L.; Calin, G.A.; Santarpia, L. Integrated MicroRNA–mRNA Profiling Identifies Oncostatin M as a Marker of Mesenchymal-Like ER-Negative/HER2-Negative Breast Cancer. Int. J. Mol. Sci. 2017, 18, 194. https://doi.org/10.3390/ijms18010194
Bottai G, Diao L, Baggerly KA, Paladini L, Győrffy B, Raschioni C, Pusztai L, Calin GA, Santarpia L. Integrated MicroRNA–mRNA Profiling Identifies Oncostatin M as a Marker of Mesenchymal-Like ER-Negative/HER2-Negative Breast Cancer. International Journal of Molecular Sciences. 2017; 18(1):194. https://doi.org/10.3390/ijms18010194
Chicago/Turabian StyleBottai, Giulia, Lixia Diao, Keith A. Baggerly, Laura Paladini, Balázs Győrffy, Carlotta Raschioni, Lajos Pusztai, George A. Calin, and Libero Santarpia. 2017. "Integrated MicroRNA–mRNA Profiling Identifies Oncostatin M as a Marker of Mesenchymal-Like ER-Negative/HER2-Negative Breast Cancer" International Journal of Molecular Sciences 18, no. 1: 194. https://doi.org/10.3390/ijms18010194
APA StyleBottai, G., Diao, L., Baggerly, K. A., Paladini, L., Győrffy, B., Raschioni, C., Pusztai, L., Calin, G. A., & Santarpia, L. (2017). Integrated MicroRNA–mRNA Profiling Identifies Oncostatin M as a Marker of Mesenchymal-Like ER-Negative/HER2-Negative Breast Cancer. International Journal of Molecular Sciences, 18(1), 194. https://doi.org/10.3390/ijms18010194