Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a
Abstract
:1. Introduction
2. Results
2.1. Identification of Soybean CCA1-Like Proteins and Their Phylogenetic Relationships
2.2. Chromosomal Localizations and Duplications of CCA1-Like Genes in Soybean
2.3. Tissue-Specific Expression Patterns of CCA1-Like Genes in Soybean
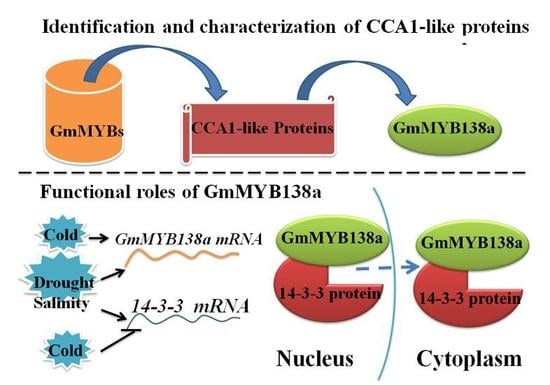
2.4. GmMYB138a Interacts with GmSGF14l
2.5. Functional Characterization of GmMYB138a
2.6. Expression Patterns of GmMYB138a and GmGF14l in Response to Abiotic Stresses
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. In silico Analysis
4.3. Multiple Sequence Alignment and Phylogenetic Analysis
4.4. Chromosomal Localization and Gene Duplication
4.5. Gene Expression Analysis
4.6. Targeted Yeast Two-Hybrid (Y2H) Assay
4.7. Bimolecular Fluorescence Complementation (BiFC) Assay
4.8. Predictions of pST-Binding Motif and Protein Interaction
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Liu, J.Y.; Osbourn, A.; Ma, P.D. MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 2015, 8, 689–708. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Du, H.; Wang, Y.B.; Xie, Y.; Liang, Z.; Jiang, S.J.; Zhang, S.S.; Huang, Y.B.; Tang, Y.X. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013, 20, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Green, R.M.; Tobin, E.M. The role of CCA1 and LHY in the plant circadian clock. Dev. Cell 2002, 2, 516–518. [Google Scholar] [CrossRef]
- Li, X.; Ma, D.B.; Lu, S.X.; Hu, X.Y.; Huang, R.F.; Liang, T.; Xu, T.D.; Tobin, E.M.; Liu, H.T. Blue light- and low temperature-regulated COR27 and COR28 play roles in the Arabidopsis circadian clock. Plant Cell 2016, 28, 2755–2769. [Google Scholar] [CrossRef] [PubMed]
- Oda, A.; Higuchi, Y.; Hisamatsu, T. Photoperiod-insensitive floral transition in chrysanthemum induced by constitutive expression of chimeric repressor CsLHY-SRDX. Plant Sci. 2017, 259, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.X.; Derynck, M.R.; Li, X.Y.; Telmer, P.; Marsolais, F.; Dhaubhadel, S. A single-repeat MYB transcription factor, GmMYB176, regulates CHS8 gene expression and affects isoflavonoid biosynthesis in soybean. Plant J. 2010, 62, 1019–1034. [Google Scholar] [CrossRef] [PubMed]
- Lai, A.G.; Doherty, C.J.; Mueller-Roeber, B.; Kay, S.A.; Schippers, J.H.M.; Dijkwel, P.P. CIRCADIAN CLOCK-ASSOCIATED 1 regulates ROS homeostasis and oxidative stress responses. Proc. Natl. Acad. Sci. USA 2012, 109, 17129–17134. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Webb, C.J.; Knowles, S.M.; Kim, S.H.J.; Wang, Z.Y.; Tobin, E.M. CCA1 and ELF3 interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 2012, 158, 1079–1088. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Kwon, Y.J.; Gil, K.E.; Park, C.M. LATE ELONGATED HYPOCOTYL regulates photoperiodic flowering via the circadian clock in Arabidopsis. BMC Plant Biol. 2016, 16, 114. [Google Scholar] [CrossRef] [PubMed]
- Penfield, S.; Hall, A. A role for multiple circadian clock genes in the response to signals that break seed dormancy in Arabidopsis. Plant Cell 2009, 21, 1722–1732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.B.; Chen, Y.H.; Wang, Z.Y.; Chen, Z.L.; Gu, H.Y.; Qu, L.J. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 2007, 51, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Nagel, D.H.; Doherty, C.J.; Pruneda-Paz, J.L.; Schmitz, R.J.; Ecker, J.R.; Kay, S.A. Genome-wide identification of CCA1 targets uncovers an expanded clock network in Arabidopsis. Proc. Natl. Acad. Sci. USA 2015, 112, E4802–E4810. [Google Scholar] [CrossRef] [PubMed]
- Perez, G.P.; Ma, Y.; Yanovsky, M.J.; Mas, P. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 5249–5253. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Jeong, C.Y.; Kang, G.H.; Yoo, S.D.; Hong, S.W.; Lee, H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J. 2015, 84, 1192–1205. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.M.; Xu, G.; Jing, Y.J.; Tang, W.J.; Lin, R.C. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 2016, 7, 12377. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.K.; Lo, P.C.; Huang, L.F.; Wu, S.J.; Yeh, C.H.; Lu, C.A. A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 2015, 88, 269–286. [Google Scholar] [CrossRef] [PubMed]
- Deng, C.Y.; Ye, H.Y.; Fan, M.; Pu, T.L.; Yan, J.B. The rice transcription factors OsICE confer enhanced cold tolerance in transgenic Arabidopsis. Plant Signal Behav. 2017, 12, e1316442. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Chao, Y.C.; Tseng, T.W.; Huang, C.K.; Lo, P.C.; Lu, C.A. Two MYB-related transcription factors play opposite roles in sugar signaling in Arabidopsis. Plant Mol. Biol. 2017, 93, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Chen, L.; Dhaubhadel, S. 14-3-3 proteins regulate the intracellular localization of the transcriptional activator GmMYB176 and affect isoflavonoid synthesis in soybean. Plant J. 2012, 71, 239–250. [Google Scholar] [CrossRef] [PubMed]
- Yao, H.Y.; Wang, G.L.; Guo, L.; Wang, X.M. Phosphatidic acid interacts with a MYB transcription factor and regulates its nuclear localization and function in Arabidopsis. Plant Cell 2013, 25, 5030–5042. [Google Scholar] [CrossRef] [PubMed]
- Frerigmann, H.; Berger, B.; Gigolashvili, T. bHLH05 is an interaction partner of MYB51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 2014, 166, 349–369. [Google Scholar] [CrossRef] [PubMed]
- Gokirmak, T.; Paul, A.L.; Ferl, R.J. Plant phosphopeptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 2010, 13, 527–532. [Google Scholar] [CrossRef] [PubMed]
- Jaspert, N.; Throm, C.; Oecking, C. Arabidopsis 14-3-3 proteins: Fascinating and less fascinating aspects. Front Plant Sci. 2011, 2, 96. [Google Scholar] [CrossRef] [PubMed]
- Denison, F.C.; Paul, A.L.; Zupanska, A.K.; Ferl, R.J. 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 2011, 22, 720–727. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Dhaubhadel, S. 14-3-3 proteins act as scaffolds for GmMYB62 and GmMYB176 and regulate their intracellular localization in soybean. Plant Signal Behav. 2012, 7, 965–968. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Y.; Dhaubhadel, S. Soybean 14-3-3 gene family: Identification and molecular characterization. Planta 2011, 233, 569–582. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Shin, J.H.; Kang, Y.J.; Shim, S.R.; Lee, S.H. Divergence of flowering genes in soybean. J. Biosci. 2012, 37, 857–870. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Wang, H.G.; Gao, P.F.; Xu, J.H.; Xu, T.D.; Wang, J.S.; Wang, B.L.; Lin, C.T.; Fu, Y.F. Analysis of clock gene homologs using unifoliolates as target organs in soybean (Glycine max). J. Plant Physiol. 2009, 166, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Zou, H.F.; Wang, H.W.; Zhang, W.K.; Ma, B.; Zhang, J.S.; Chen, S.Y. Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res. 2008, 18, 1047–1060. [Google Scholar] [CrossRef] [PubMed]
- Klug, A. Zinc finger peptides for the regulation of gene expression. J. Mol. Biol. 1999, 293, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.S. Zinc finger proteins: Getting a grip on RNA. Curr. Opin. Struc. Biol. 2005, 15, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Leister, D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004, 20, 116–122. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Zhao, Y.; Cao, J.; Zhang, W.; Jiang, H.; Li, X.; Ma, Q.; Zhu, S.; Cheng, B. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 2012, 7, e40120. [Google Scholar] [CrossRef] [PubMed]
- Bian, S.M.; Li, X.Y.; Mainali, H.; Chen, L.; Dhaubhadel, S. Genome-wide analysis of DWD proteins in soybean (Glycine max): Significance of Gm08DWD and GmMYB176 interaction in isoflavonoid biosynthesis. PLoS ONE 2017, 12, e0178947. [Google Scholar] [CrossRef] [PubMed]
- Prouse, M.B.; Campbell, M.M. The interaction between MYB proteins and their target DNA binding sites. Biochim. Biophys. Acta 2012, 1819, 67–77. [Google Scholar] [CrossRef] [PubMed]
- Lavin, M.; Herendeen, P.S.; Wojciechowski, M.F. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005, 54, 575–594. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.X.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.J.; Thelen, J.J.; Cheng, J.L.; et al. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Kim, J.H.; Nguyen, H.N.; Jikumaru, Y.; Kamiya, Y.; Hong, S.W.; Lee, H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 2013, 64, 3911–3922. [Google Scholar] [CrossRef] [PubMed]
- Lu, D.; Wang, T.; Persson, S.; Mueller-Roeber, B.; Schippers, J.H.M. Transcriptional control of ROS homeostasis by KUODA1 regulates cell expansion during leaf development. Nat. Commun. 2014. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.A.; Shalit-Kaneh, A.; Chu, D.N.; Hsu, P.Y.; Harmer, S.L. The REVEILLE clock genes inhibit growth of juvenile and adult plants by control of cell size. Plant Physiol. 2017, 173, 2308–2322. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, M.J.; Lim, M.H.; Kim, S.G.; Lee, M.; Baldwin, I.T.; Park, C.M. A self-regulatory circuit of CIRCADIAN CLOCK-ASSOCIATED1 underlies the circadian clock regulation of temperature responses in Arabidopsis. Plant Cell 2012, 24, 2427–2442. [Google Scholar] [CrossRef] [PubMed]
- Marshall, C.M.; Tartaglio, V.; Duarte, M.; Harmon, F.G. The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 2016, 28, 2560–2575. [Google Scholar] [CrossRef] [PubMed]
- Filichkin, S.A.; Mockler, T.C. Unproductive alternative splicing and nonsense mRNAs: A widespread phenomenon among plant circadian clock genes. Biol. Direct. 2012, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.; Bendix, C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; de Oliveira, M.C.N.; Harmon, F.G.; Nepomuceno, A. Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS ONE 2014, 9, e864. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Shin, J.; Davis, S.J. Abiotic stress and the plant circadian clock. Plant Signal Behav. 2011, 6, 223–231. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Luo, X.; Sun, M.Z.; Chen, C.; Ding, X.D.; Wang, X.D.; Yang, S.S.; Yu, Q.Y.; Jia, B.W.; Ji, W.; et al. A Glycine soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 2014, 55, 99–118. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zhang, Y.; Chen, L.H.; Wu, C.L.; Luo, Q.C.; Zhang, F.; Wei, Q.H.; Li, K.X.; Chang, J.L.; Yang, G.X.; He, G.Y. A member of the 14-3-3 gene family in brachypodium distachyon, BdGF14d, confers salt tolerance in transgenic tobacco plants. Front Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Sun, G.L.; Xie, F.L.; Zhang, B.H. Transcriptome-wide identification and stress properties of the 14-3-3 gene family in cotton (Gossypium hirsutum L.). Funct. Integr. Genom. 2011, 11, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Catala, R.; Lopez-Cobollo, R.; Castellano, M.M.; Angosto, T.; Alonso, J.M.; Ecker, J.R.; Salinas, J. The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 2014, 26, 3326–3342. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.Y.; Jia, Y.X.; Ding, Y.L.; Shi, Y.T.; Li, Z.; Guo, Y.; Gong, Z.Z.; Yang, S.H. Plasma membrane CRPK1-mediated phosphorylation of 14-3-3 proteins induces their nuclear import to fine-tune CBF signaling during cold response. Mol. Cell 2017, 66, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Li, R.H.; Jiang, X.T.; Jin, D.H.; Dhaubhadel, S.; Bian, S.M.; Li, X.Y. Identification of 14-3-3 family in common bean and their response to abiotic stress. PLoS ONE 2015, 10, e0143280. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xie, X.; Li, J.; Cui, Y.; Hou, Y.; Zhai, L.; Wang, X.; Fu, Y.; Liu, R.; Bian, S. Conservation and diversification of the miR166 family in soybean and potential roles of newly identified miR166s. BMC Plant Biol. 2017, 17, 32. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.K.; Wang, Y.B.; Liu, Z.X.; Cheng, H.; Xue, Y. HemI: A toolkit for illustrating heatmaps. PLoS ONE 2014, 9, e111988. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Tang, X.R.; Tian, G.; Wang, F.; Liu, K.D.; Nguyen, V.; Kohalmi, S.E.; Keller, W.A.; Tsang, E.W.T.; Harada, J.J.; Rothstein, S.J.; Cui, Y.H. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J. 2010, 61, 259–270. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bian, S.; Jin, D.; Li, R.; Xie, X.; Gao, G.; Sun, W.; Li, Y.; Zhai, L.; Li, X. Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a. Int. J. Mol. Sci. 2017, 18, 2040. https://doi.org/10.3390/ijms18102040
Bian S, Jin D, Li R, Xie X, Gao G, Sun W, Li Y, Zhai L, Li X. Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a. International Journal of Molecular Sciences. 2017; 18(10):2040. https://doi.org/10.3390/ijms18102040
Chicago/Turabian StyleBian, Shaomin, Donghao Jin, Ruihua Li, Xin Xie, Guoli Gao, Weikang Sun, Yuejia Li, Lulu Zhai, and Xuyan Li. 2017. "Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a" International Journal of Molecular Sciences 18, no. 10: 2040. https://doi.org/10.3390/ijms18102040
APA StyleBian, S., Jin, D., Li, R., Xie, X., Gao, G., Sun, W., Li, Y., Zhai, L., & Li, X. (2017). Genome-Wide Analysis of CCA1-Like Proteins in Soybean and Functional Characterization of GmMYB138a. International Journal of Molecular Sciences, 18(10), 2040. https://doi.org/10.3390/ijms18102040