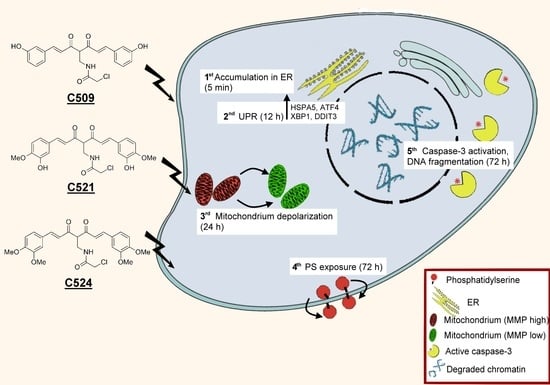
Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells
Abstract
:1. Introduction
2. Results and Discussion
2.1. Achiral Curcumin Analogs Impaired Tumor Cell Viability
2.2. Curcumin Analogs Induced Phosphatidylserine Exposure of A549, HepG2 and PANC-1 Cells
2.3. Curcumin Analogs Caused G0/G1 Cell-Cycle Arrest of PANC-1 Cells
2.4. Curcumin Analogs Induced ER (Endoplasmic Reticulum) Stress and Mitochondrial Membrane Depolarization
2.5. Curcumin Analogs Induced Caspase-3 Activation and DNA Fragmentation
3. Materials and Methods
3.1. Cell Culturing and Treatments
3.2. Resazurin Viability Assay
3.3. Flow Cytometry
3.3.1. Detection of Phosphatidylserine Exposure
3.3.2. Cell Cycle and Sub-G1 Analysis
3.3.3. Immunofluorescence
3.3.4. Detection of the Loss of Mitochondrial Membrane Potential
3.4. Confocal Laser Scanning Microscopy
3.5. Gene Expression Analysis
3.6. Statistical Analysis
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| AnnV | Annexin V-Alexa Fluor® 488 |
| CDKN2A | Cyclin-dependent kinase inhibitor 2A |
| CDK4 | Cyclin-dependent kinase 4 |
| CDK6 | Cyclin-dependent kinase 6 |
| Curc | Curcumin |
| ER | Endoplasmic reticulum |
| GLUT1 | Glucose transporter-1 |
| GRAS | Generally Recognized as Safe |
| HCC | Hepatocellular carcinoma |
| HEPES | 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid |
| IC50 | Half maximal inhibitory concentration |
| JC-1 | 5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethylbenzimidazolocarbocyanine iodide |
| NSCLC | Non-small cell lung cancer |
| MAPK | Mitogen-activated protein kinase |
| MFI | Median fluorescence intensity |
| MMP | Mitochondrial membrane potential |
| PBS | Phosphate buffered saline |
| PI | Propidium iodide |
| SAR | structure-activity relationship |
| SD | Standard deviation |
| UPR | Unfolded protein response |
References
- Gupta, S.C.; Sung, B.; Kim, J.H.; Prasad, S.; Li, S.; Aggarwal, B.B. Multitargeting by turmeric, the golden spice: From kitchen to clinic. Mol. Nutr. Food Res. 2013, 57, 1510–1528. [Google Scholar] [CrossRef] [PubMed]
- Kocaadam, B.; Sanlier, N. Curcumin, an active component of turmeric (Curcuma longa), and its effects on health. Crit. Rev. Food Sci. Nutr. 2017, 57, 2889–2895. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; He, Z.M.; Wang, F.L.; Zhang, Z.S.; Liu, X.Z.; Zhai, D.D.; Chen, W.D. Curcumin and its promise as an anticancer drug: An analysis of its anticancer and antifungal effects in cancer and associated complications from invasive fungal infections. Eur. J. Pharmacol. 2016, 772, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Ravindran, J.; Prasad, S.; Aggarwal, B.B. Curcumin and cancer cells: How many ways can curry kill tumor cells selectively? AAPS J. 2009, 11, 495–510. [Google Scholar] [CrossRef] [PubMed]
- Perrone, D.; Ardito, F.; Giannatempo, G.; Dioguardi, M.; Troiano, G.; Lo Russo, L.; de Lillo, A.; Laino, L.; Lo Muzio, L. Biological and therapeutic activities, and anticancer properties of curcumin. Exp. Ther. Med. 2015, 10, 1615–1623. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Qiao, F.; Wang, Y.; Xu, Y.; Shang, Y. Curcumin inhibits cell proliferation and induces apoptosis of human non-small cell lung cancer cells through the upregulation of mir-192-5p and suppression of PI3K/Akt signaling pathway. Oncol. Rep. 2015, 34, 2782–2789. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Lin, M.; Wang, Y.; Lai, Y.; Hu, J.; Fu, T.; Wang, L.; Lin, S.; Chen, L.; Guo, Y. Curcumin induces the apoptosis of a549 cells via oxidative stress and MAPK signaling pathways. Int. J. Mol. Med. 2015, 36, 1118–1126. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.R.; Liu, P.L.; Chen, Y.H.; Chou, S.H.; Cheng, Y.J.; Hwang, J.J.; Chong, I.W. Curcumin inhibits non-small cell lung cancer cells metastasis through the adiponectin/nf-kappab/mmps signaling pathway. PLoS ONE 2015, 10, e0144462. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Wang, Z.; Deng, Z.; Ren, H.; Li, X. Curcumin inhibits lung cancer invasion and metastasis by attenuating glut1/mt1-mmp/mmp2 pathway. Int. J. Clin. Exp. Med. 2015, 8, 8948–8957. [Google Scholar] [PubMed]
- Wang, M.; Ruan, Y.; Chen, Q.; Li, S.; Wang, Q.; Cai, J. Curcumin induced hepg2 cell apoptosis-associated mitochondrial membrane potential and intracellular free ca(2+) concentration. Eur. J. Pharmacol. 2011, 650, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Tian, W.; Ma, X. Curcumin induces apoptosis of hepg2 cells via inhibiting fatty acid synthase. Target. Oncol. 2014, 9, 279–286. [Google Scholar] [CrossRef] [PubMed]
- Ramos, M.C.; Boulaiz, H.; Grinan-Lison, C.; Marchal, J.A.; Vicente, F. What’s new in treatment of pancreatic cancer: A patent review (2010–2017). Expert Opin. Ther. Pat. 2017. [Google Scholar] [CrossRef] [PubMed]
- Diaz Osterman, C.J.; Gonda, A.; Stiff, T.; Sigaran, U.; Valenzuela, M.M.; Ferguson Bennit, H.R.; Moyron, R.B.; Khan, S.; Wall, N.R. Curcumin induces pancreatic adenocarcinoma cell death via reduction of the inhibitors of apoptosis. Pancreas 2016, 45, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Li, C.; Xi, H.; Gao, Y.; Xu, D. Curcumin induces apoptosis in pancreatic cancer cells through the induction of forkhead box o1 and inhibition of the PI3K/Akt pathway. Mol. Med. Rep. 2015, 12, 5415–5422. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.I.; Feher, L.Z.; Szebeni, G.J.; Gyuris, M.; Sipos, P.; Alfoldi, R.; Ozsvari, B.; Hackler, L., Jr.; Balazs, A.; Batar, P.; et al. Curcumin and its analogue induce apoptosis in leukemia cells and have additive effects with bortezomib in cellular and xenograft models. Biomed. Res. Int. 2015. [Google Scholar] [CrossRef] [PubMed]
- Hackler, L., Jr.; Ozsvari, B.; Gyuris, M.; Sipos, P.; Fabian, G.; Molnar, E.; Marton, A.; Farago, N.; Mihaly, J.; Nagy, L.I.; et al. The curcumin analog C-150, influencing NF-kB, UPR and Akt/Notch pathways has potent anticancer activity in vitro and in vivo. PLoS ONE 2016, 11, e0149832. [Google Scholar] [CrossRef] [PubMed]
- Gyuris, M.; Hackler, L., Jr.; Nagy, L.I.; Alfoldi, R.; Redei, E.; Marton, A.; Vellai, T.; Farago, N.; Ozsvari, B.; Hetenyi, A.; et al. Mannich curcuminoids as potent anticancer agents. Arch. Pharm. 2017, 350, e1700005. [Google Scholar] [CrossRef] [PubMed]
- Gyuris, M.; Puskas, L.G.; Kanizsai, I.; Ozsvari, B.; Hackler, L.; Nagy, L.I. Novel Medicinal Compounds—Preparation of Curcumin Derivatives for Treatment of Cancer and Inflammation. International Patent Application No. PCT WO2013041895 A1, 28 March 2013. [Google Scholar]
- Jackson, S.J.; Murphy, L.L.; Venema, R.C.; Singletary, K.W.; Young, A.J. Curcumin binds tubulin, induces mitotic catastrophe, and impedes normal endothelial cell proliferation. Food Chem. Toxicol. 2013, 60, 431–438. [Google Scholar] [CrossRef] [PubMed]
- Hu, A.; Huang, J.J.; Zhang, J.F.; Dai, W.J.; Li, R.L.; Lu, Z.Y.; Duan, J.L.; Li, J.P.; Chen, X.P.; Fan, J.P.; et al. Curcumin induces G2/M cell cycle arrest and apoptosis of head and neck squamous cell carcinoma in vitro and in vivo through ATM/Chk2/p53-dependent pathway. Oncotarget 2017, 8, 50747–50760. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.K.; Chen, Q.; Siddiqui, I.; Sarva, K.; Shankar, S. Linkage of curcumin-induced cell cycle arrest and apoptosis by cyclin-dependent kinase inhibitor p21(/waf1/cip1). Cell Cycle 2007, 6, 2953–2961. [Google Scholar] [CrossRef] [PubMed]
- Sha, J.; Li, J.; Wang, W.; Pan, L.; Cheng, J.; Li, L.; Zhao, H.; Lin, W. Curcumin induces g0/g1 arrest and apoptosis in hormone independent prostate cancer du-145 cells by down regulating notch signaling. Biomed. Pharmacother. 2016, 84, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Nagy, L.I.; Molnar, E.; Kanizsai, I.; Madacsi, R.; Ozsvari, B.; Feher, L.Z.; Fabian, G.; Marton, A.; Vizler, C.; Ayaydin, F.; et al. Lipid droplet binding thalidomide analogs activate endoplasmic reticulum stress and suppress hepatocellular carcinoma in a chemically induced transgenic mouse model. Lipids Health Dis. 2013, 12, 175. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.; Kim, H.S.; Jung, E.J.; Lee, J.Y.; Tsang, B.K.; Lim, J.M.; Song, Y.S. Curcumin induces ER stress-mediated apoptosis through selective generation of reactive oxygen species in cervical cancer cells. Mol. Carcinog. 2016, 55, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Chignell, C.F.; Bilski, P.; Reszka, K.J.; Motten, A.G.; Sik, R.H.; Dahl, T.A. Spectral and photochemical properties of curcumin. Photochem. Photobiol. 1994, 59, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Gessner, D.K.; Schlegel, G.; Ringseis, R.; Schwarz, F.J.; Eder, K. Up-regulation of endoplasmic reticulum stress induced genes of the unfolded protein response in the liver of periparturient dairy cows. BMC Vet. Res. 2014, 10, 46. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, M.; Rocchi, S. New anti-cancer molecules targeting HSPA5/BIP to induce endoplasmic reticulum stress, autophagy and apoptosis. Autophagy 2017, 13, 216–217. [Google Scholar] [CrossRef] [PubMed]
- Cnop, M.; Foufelle, F.; Velloso, L.A. Endoplasmic reticulum stress, obesity and diabetes. Trends Mol. Med. 2012, 18, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Guo, Y.; Tang, J.; Jiang, J.; Chen, Z. New insights into the roles of chop-induced apoptosis in ER stress. Acta Biochim. Biophys. Sin. 2014, 46, 629–640. [Google Scholar] [CrossRef] [PubMed]
- Sano, R.; Reed, J.C. Er stress-induced cell death mechanisms. Biochim. Biophys. Acta 2013, 1833, 3460–3470. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.; Cuffe, L.; Szegezdi, E.; Logue, S.E.; Neary, C.; Healy, S.; Samali, A. Mechanisms of ER stress-mediated mitochondrial membrane permeabilization. Int. J. Cell Biol. 2010, 2010, 170215. [Google Scholar] [CrossRef] [PubMed]
- Henry-Mowatt, J.; Dive, C.; Martinou, J.C.; James, D. Role of mitochondrial membrane permeabilization in apoptosis and cancer. Oncogene 2004, 23, 2850–2860. [Google Scholar] [CrossRef] [PubMed]
- Kajstura, M.; Halicka, H.D.; Pryjma, J.; Darzynkiewicz, Z. Discontinuous fragmentation of nuclear DNA during apoptosis revealed by discrete “sub-g1” peaks on DNA content histograms. Cytometry A 2007, 71, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Duellman, S.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. Available online: http://www.ncbi.nlm.nih.gov/pubmed/23805433 (accessed on 26 September 2017).
- Fajka-Boja, R.; Hidvegi, M.; Shoenfeld, Y.; Ion, G.; Demydenko, D.; Tomoskozi-Farkas, R.; Vizler, C.; Telekes, A.; Resetar, A.; Monostori, E. Fermented wheat germ extract induces apoptosis and downregulation of major histocompatibility complex class I proteins in tumor T and B cell lines. Int. J. Oncol. 2002, 20, 563–570. [Google Scholar] [CrossRef] [PubMed]
- Sharrow, S.O. Analysis of flow cytometry data. Curr. Protoc. Immunol. 1996. [Google Scholar] [CrossRef]
- Puskas, L.G.; Feher, L.Z.; Vizler, C.; Ayaydin, F.; Raso, E.; Molnar, E.; Magyary, I.; Kanizsai, I.; Gyuris, M.; Madacsi, R.; et al. Polyunsaturated fatty acids synergize with lipid droplet binding thalidomide analogs to induce oxidative stress in cancer cells. Lipids Health Dis. 2010, 9, 56. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, G.J.; Tancos, Z.; Feher, L.Z.; Alfoldi, R.; Kobolak, J.; Dinnyes, A.; Puskas, L.G. Real architecture for 3D tissue (raft) culture system improves viability and maintains insulin and glucagon production of mouse pancreatic islet cells. Cytotechnology 2017, 69, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Hughes, P.E.; Caenepeel, S.; Wu, L.C. Targeted therapy and checkpoint immunotherapy combinations for the treatment of cancer. Trends Immunol. 2016, 37, 462–476. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Teicher, B.A.; Hassan, R. Antibody-drug conjugates for cancer therapy. Lancet Oncol. 2016, 17, e254–e262. [Google Scholar] [CrossRef]
- Al-Lazikani, B.; Banerji, U.; Workman, P. Combinatorial drug therapy for cancer in the post-genomic era. Nat. Biotechnol. 2012, 30, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Szebeni, G.J.; Vizler, C.; Nagy, L.I.; Kitajka, K.; Puskas, L.G. Pro-tumoral inflammatory myeloid cells as emerging therapeutic targets. Int. J. Mol. Sci. 2016, 17, 1958. [Google Scholar] [CrossRef] [PubMed]









| Compound | A549 | HepG2 | PANC-1 |
|---|---|---|---|
| Curc. | 27.11 | 14.53 | 30.57 |
| C501 # | 1.26 | 0.66 | 1.44 |
| C502 | 2.39 | >5 | 2.20 |
| C503 # | 1.70 | 2.55 | 1.66 |
| C504 # | 1.58 | 1.24 | 1.40 |
| C505 # | 1.29 | 1.48 | 1.43 |
| C509 # | 2.27 | 2.98 | 1.01 |
| C510 | >5 | 1.07 | >5 |
| C513 | >5 | 3.11 | >5 |
| C514 | inactive | inactive | inactive |
| C515 # | 2.73 | 1.31 | 1.46 |
| C516 | >5 | 4.19 | >5 |
| C517 | >5 | >5 | 2.45 |
| C518 # | 2.42 | 4.77 | 1.25 |
| C519 | >5 | >5 | >5 |
| C520 | inactive | inactive | inactive |
| C521 # | 1.88 | 1.36 | 1.18 |
| C524 # | 1.70 | 1.69 | 1.14 |
| C525 | inactive | inactive | inactive |
| C526 | inactive | inactive | inactive |
| C529 | inactive | inactive | inactive |
| C530 | inactive | inactive | inactive |
| C532 | >5 | >5 | 1.58 |
| C533 | >5 | 4.81 | >5 |
| Compound | A549 | HepG2 | PANC-1 |
|---|---|---|---|
| None | 2.4 ± 0.44 | 2.7 ± 0.37 | 1.8 ± 0.02 |
| Curcumin | 3.0 ± 0.30 | 2.0 ± 0.14 | 7.0 ± 0.02 |
| C501 # | 1.3 ± 0.06 | 2.6 ± 0.55 | 6.3 ± 0.42 |
| C503 | 1.9 ± 0.83 | 2.6 ± 0.33 | 6.1 ± 0.54 |
| C504 | 2.0 ± 0.16 | 3.4 ± 0.28 | 8.8 ± 1.36 |
| C505 | 1.8 ± 0.11 | 5.6 ± 0.31 | 8.2 ± 5.83 |
| C509 # | 2.7 ± 0.25 | 15.4 ± 9.92 | 51.0 ± 1.17 |
| C515 | 3.0 ± 0.77 | 3.1 ± 0.11 | 22.5 ± 4.43 |
| C518 | 3.0 ± 0.01 | 3.6 ± 0.33 | 11.8 ± 2.49 |
| C521 # | 2.5 ± 0.5 | 8.9 ± 0.13 | 37.8 ± 10.86 |
| C524 # | 19.8 ± 3.81 | 7.3 ± 0.37 | 80.4 ± 0.13 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Szebeni, G.J.; Balázs, Á.; Madarász, I.; Pócz, G.; Ayaydin, F.; Kanizsai, I.; Fajka-Boja, R.; Alföldi, R.; Hackler Jr., L.; Puskás, L.G. Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells. Int. J. Mol. Sci. 2017, 18, 2105. https://doi.org/10.3390/ijms18102105
Szebeni GJ, Balázs Á, Madarász I, Pócz G, Ayaydin F, Kanizsai I, Fajka-Boja R, Alföldi R, Hackler Jr. L, Puskás LG. Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells. International Journal of Molecular Sciences. 2017; 18(10):2105. https://doi.org/10.3390/ijms18102105
Chicago/Turabian StyleSzebeni, Gábor J., Árpád Balázs, Ildikó Madarász, Gábor Pócz, Ferhan Ayaydin, Iván Kanizsai, Roberta Fajka-Boja, Róbert Alföldi, László Hackler Jr., and László G. Puskás. 2017. "Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells" International Journal of Molecular Sciences 18, no. 10: 2105. https://doi.org/10.3390/ijms18102105
APA StyleSzebeni, G. J., Balázs, Á., Madarász, I., Pócz, G., Ayaydin, F., Kanizsai, I., Fajka-Boja, R., Alföldi, R., Hackler Jr., L., & Puskás, L. G. (2017). Achiral Mannich-Base Curcumin Analogs Induce Unfolded Protein Response and Mitochondrial Membrane Depolarization in PANC-1 Cells. International Journal of Molecular Sciences, 18(10), 2105. https://doi.org/10.3390/ijms18102105