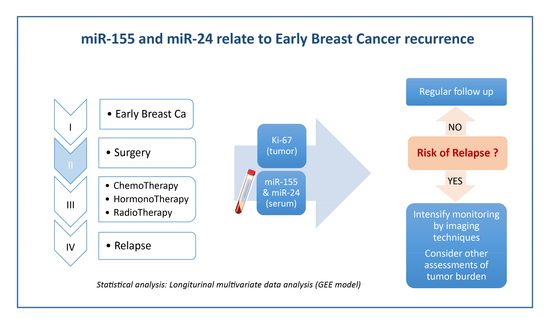
Prediction Potential of Serum miR-155 and miR-24 for Relapsing Early Breast Cancer
Abstract
:1. Introduction
2. Results
2.1. OncomiRs miR-155 and miR-24 Are Predictive of Early Breast Cancer (EBC) Relapse
2.2. Ki-67 Expression Specified the Relapse Probability Defined by Levels of miR-155 or miR-24
- a.
- Only miR-155 level is available (or considered)
- b.
- Only miR-24 level is available
- c.
- Both miR-155 and miR-24 levels are available
3. Discussion
4. Materials and Methods
4.1. Patients
4.2. Ethics Approval and Consent to Participate
4.3. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mulrane, L.; Klinger, R.; McGee, S.F.; Gallagher, W.M.; O’Connor, D.P. microRNAs: A new class of breast cancer biomarkers. Expert Rev. Mol. Diagn. 2014, 14, 347–363. [Google Scholar] [CrossRef] [PubMed]
- Zoon, C.K.; Starker, E.Q.; Wilson, A.M.; Emmert-Buck, M.R.; Libutti, S.K.; Tangrea, M.A. Current molecular diagnostics of breast cancer and the potential incorporation of microRNA. Expert Rev. Mol. Diagn. 2009, 9, 455–467. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.V.; Ferracin, M.; Liu, C.G.; Veronese, A.; Spizzo, R.; Sabbioni, S.; Magri, E.; Pedriali, M.; Fabbri, M.; Campiglio, M.; et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005, 65, 7065–7070. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Chadalapaka, G.; Lee, S.O.; Yamada, D.; Sastre-Garau, X.; Defossez, P.A.; Park, Y.Y.; Lee, J.S.; Safe, S. Identification of oncogenic microRNA-17–92/ZBTB4/specificity protein axis in breast cancer. Oncogene 2012, 31, 1034–1044. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yang, C.; Zhai, L.; Zhang, W.; Yu, J.; Gu, F.; Lang, R.; Fan, Y.; Gong, M.; Zhang, X.; et al. Deep sequencing reveals small RNA characterization of invasive micropapillary carcinomas of the breast. Breast Cancer Res. Treat. 2012, 136, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Wang, C.; Lu, Z.; Guo, L.; Ge, Q. Analysis of serum genome-wide microRNAs for breast cancer detection. Clin. Chim. Acta 2012, 413, 1058–1065. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.J.; Shin, J.Y.; Lee, K.D.; Bae, Y.K.; Sung, K.W.; Nam, S.J.; Chun, K.H. MicroRNA let-7a suppresses breast cancer cell migration and invasion through downregulation of C–C chemokine receptor type 7. Breast Cancer Res. 2012, 14, R14. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Bai, Z.; Han, W.; Zhang, J.; Meng, H.; Bi, J.; Ma, X.; Han, S.; Zhang, Z. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig. Dis. Sci. 2012, 57, 897–904. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Shen, J.; Medico, L.; Wang, D.; Ambrosone, C.B.; Liu, S. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS ONE 2010, 5, e13735. [Google Scholar] [CrossRef] [PubMed]
- Sochor, M.; Basova, P.; Pesta, M.; Dusilkova, N.; Bartos, J.; Burda, P.; Pospisil, V.; Stopka, T. Oncogenic microRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014, 14, 448. [Google Scholar] [CrossRef] [PubMed]
- Eichelser, C.; Flesch-Janys, D.; Chang-Claude, J.; Pantel, K.; Schwarzenbach, H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin. Chem. 2013, 59, 1489–1496. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; He, L.; Richards, E.J.; Challa, S.; Xu, C.X.; Permuth-Wey, J.; Lancaster, J.M.; Coppola, D.; Sellers, T.A.; Djeu, J.Y.; et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene 2014, 33, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Gasparini, P.; Lovat, F.; Fassan, M.; Casadei, L.; Cascione, L.; Jacob, N.K.; Carasi, S.; Palmieri, D.; Costinean, S.; Shapiro, C.L.; et al. Protective role of miR-155 in breast cancer through RAD51 targeting impairs homologous recombination after irradiation. Proc. Natl. Acad. Sci. USA 2014, 111, 4536–4541. [Google Scholar] [CrossRef] [PubMed]
- Bacci, M.; Giannoni, E.; Fearns, A.; Ribas, R.; Gao, Q.; Taddei, M.L.; Pintus, G.; Dowsett, M.; Isacke, C.M.; Martin, L.A.; et al. miR-155 drives metabolic reprogramming of ER+ breast cancer cells following long-term estrogen deprivation and predicts clinical response to aromatase inhibitors. Cancer Res. 2016, 76, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- Ell, B.; Qiu, Q.; Wei, Y.; Mercatali, L.; Ibrahim, T.; Amadori, D.; Kang, Y. The microRNA-23b/27b/24 cluster promotes breast cancer lung metastasis by targeting metastasis-suppressive gene prosaposin. J. Biol. Chem. 2014, 289, 21888–21895. [Google Scholar] [CrossRef] [PubMed]
- Farazi, T.A.; Horlings, H.M.; Ten Hoeve, J.J.; Mihailovic, A.; Halfwerk, H.; Morozov, P.; Brown, M.; Hafner, M.; Reyal, F.; van Kouwenhove, M.; et al. MicroRNA sequence and expression analysis in breast tumors by deep sequencing. Cancer Res. 2011, 71, 4443–4453. [Google Scholar] [CrossRef] [PubMed]
- Bisso, A.; Faleschini, M.; Zampa, F.; Capaci, V.; De Santa, J.; Santarpia, L.; Piazza, S.; Cappelletti, V.; Daidone, M.; Agami, R.; et al. Oncogenic miR-181a/b affect the DNA damage response in aggressive breast cancer. Cell Cycle 2013, 12, 1679–1687. [Google Scholar] [CrossRef] [PubMed]
- Serpico, D.; Molino, L.; Di Cosimo, S. microRNAs in breast cancer development and treatment. Cancer Treat. Rev. 2014, 40, 595–604. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Zeger, S.L. Longitudinal data analysis using generalized linear models. Biometrika 1986, 73, 13–22. [Google Scholar] [CrossRef]
- Hudecova, S.; Pesta, M. Modeling dependencies in claims reserving with GEE. Insur. Math. Econ. 2013, 53, 786–794. [Google Scholar] [CrossRef]


| Risk Characteristic | Estimate | Standard Error | p-Value | |
|---|---|---|---|---|
| (A) | Intercept | 4.019 | 0.715 | <0.001 |
| miR-155 | −0.428 | 0.194 | 0.028 | |
| Ki67 ≥ 20% | −1.788 | 0.717 | 0.013 | |
| Triple negative | −0.093 | 1.937 | 0.961 | |
| HER2 positive | −0.536 | 1.865 | 0.774 | |
| Grade III | −0.960 | 0.897 | 0.284 | |
| Node positive | −0.113 | 1.035 | 0.913 | |
| Chemotherapy (yes) | −1.738 | 1.163 | 0.135 | |
| Radiotherapy (yes) | 1.600 | 1.188 | 0.178 | |
| Hormonal therapy (yes) | 0.936 | 1.674 | 0.576 | |
| Family history (yes) | −0.407 | 1.197 | 0.734 | |
| (B) | Intercept | 5.023 | 1.038 | <0.001 |
| miR-24 | −1.311 | 0.565 | 0.020 | |
| Ki67 ≥ 20% | −1.633 | 0.719 | 0.023 | |
| Triple negative | 1.444 | 2.287 | 0.528 | |
| HER2 positive | −0.030 | 2.047 | 0.988 | |
| Grade III | −0.991 | 0.875 | 0.258 | |
| Node positive | −0.593 | 1.007 | 0.556 | |
| Chemotherapy (yes) | −2.364 | 1.239 | 0.056 | |
| Radiotherapy (yes) | 1.526 | 1.174 | 0.194 | |
| Hormonal therapy (yes) | 1.682 | 1.968 | 0.393 | |
| Family history (yes) | −0.177 | 1.161 | 0.879 | |
| (C) | Intercept | 5.687 | 1.179 | <0.001 |
| miR-155 | −0.396 | 0.190 | 0.038 | |
| miR-24 | −1.206 | 0.591 | 0.041 | |
| Ki67 ≥ 20% | −1.605 | 0.743 | 0.031 | |
| Triple negative | 0.681 | 2.331 | 0.770 | |
| HER2 positive | −0.282 | 2.028 | 0.889 | |
| Grade III | −1.197 | 0.949 | 0.207 | |
| Node positive | −0.369 | 1.117 | 0.741 | |
| Chemotherapy (yes) | −3.005 | 1.549 | 0.052 | |
| Radiotherapy (yes) | 1.709 | 1.337 | 0.201 | |
| Hormonal therapy (yes) | 1.298 | 1.939 | 0.503 | |
| Family history (yes) | −0.109 | 1.269 | 0.932 |
| Clinical Parameters | Values |
|---|---|
| Number of patients | 133 |
| Number of tumors | 134 |
| Age | Median 61.5 (37–84) years |
| Follow-up | Median 53.25 (21.5–68.5) months |
| PFS (progression free survival) | Median 51.5 (11–67.5) months |
| Deaths due to cancer (overall deaths) | 2 (6) |
| premenopausal | 23 |
| postmenopausal | 110 |
| Histology ductal | 97 (72%) |
| Histology lobular | 10 (8%) |
| Histology mixed + others | 27 (20%) |
| Tumors in personal history | 15 (11%) |
| pT1a (tumor 0.1–0.5 cm) | 3 (2%) |
| pT1b (tumor 0.5–1.0 cm) | 21 (16%) |
| pT1c (tumor 1.0–2.0 cm) | 70 (52%) |
| pT2 (tumor 2.0–5.0 cm) | 40 (30%) |
| pT3 (tumor > 5 cm) | 0 |
| pT4 (invasion to chest, skin, inflam. BC) | 0 |
| N+ (positive lymphatic nodes) | 26 (19%) |
| N− (negative lymphatic nodes) | 108 (81%) |
| G I (histological grade 1) | 27 (20%) |
| G II (histological grade 2) | 72 (54%) |
| G III (histological grade 3) | 27 (20%) |
| No grade | 8 (6%) |
| HR+ (positive for hormonal receptors) | 119 (89%) |
| HR− (negative for hormonal receptors) | 15 (11%) |
| HER+ (HER2 positive) | 4 (3%) |
| HER− (HER2 negative) | 125 (93%) |
| No HER | 5 (4%) |
| Ki-67 0–19% of positive cells | 100 (75%) |
| Ki-67 > 20% of positive cells | 29 (21%) |
| No Ki-67 (negative) | 5 (4%) |
| Triple negative (HR/HER2 negativity) | 12 (9%) |
| Low-risk group | 65 (49%) |
| High-risk group | 68 (51%) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bašová, P.; Pešta, M.; Sochor, M.; Stopka, T. Prediction Potential of Serum miR-155 and miR-24 for Relapsing Early Breast Cancer. Int. J. Mol. Sci. 2017, 18, 2116. https://doi.org/10.3390/ijms18102116
Bašová P, Pešta M, Sochor M, Stopka T. Prediction Potential of Serum miR-155 and miR-24 for Relapsing Early Breast Cancer. International Journal of Molecular Sciences. 2017; 18(10):2116. https://doi.org/10.3390/ijms18102116
Chicago/Turabian StyleBašová, Petra, Michal Pešta, Marek Sochor, and Tomáš Stopka. 2017. "Prediction Potential of Serum miR-155 and miR-24 for Relapsing Early Breast Cancer" International Journal of Molecular Sciences 18, no. 10: 2116. https://doi.org/10.3390/ijms18102116
APA StyleBašová, P., Pešta, M., Sochor, M., & Stopka, T. (2017). Prediction Potential of Serum miR-155 and miR-24 for Relapsing Early Breast Cancer. International Journal of Molecular Sciences, 18(10), 2116. https://doi.org/10.3390/ijms18102116