Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis
Abstract
:1. Introduction
2. Results
2.1. Changes of Photosynthetic Performance in WHY1 Mutants
2.2. WHY1 Affects the Expression of PSI-LHCI Encoding Genes
2.3. WHY1 Interacts with LHCA1 In Vitro and In Vivo
2.4. WHY1 Affects the Expression of Plastid NDH Genes
2.5. NDH Activity and the Accumulation of NDH18 Were Changed in WHY1 Mutants
2.6. WHIRLY Proteins Alter the Photosynthetic Performances after High Light Treatment
2.7. WHIRLY Proteins Affect the Expression of Photosynthesis Related Genes under High Light Condition
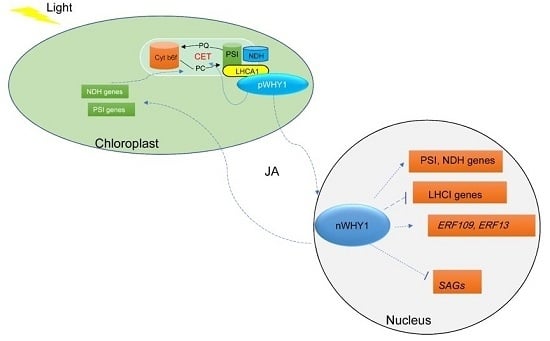
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Analysis of Photosynthetic Parameters
4.3. Quantitative Real-Time PCR
4.4. Yeast Two-Hybrid System Screen and Confirmation
4.5. Immunological Analyses
4.6. Coimmunoprecipitation of Photosystem I Complex Proteins
4.7. Bimolecular Fluorescence Complementation Assay (BiFC)
4.8. In Vivo Measurements of NDH Activity
4.9. Separation of Photosynthetic Complexes by Blue-Native Polyacrylamide Gel Electrophoresis (BN-PAGE)
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Pfannschmidt, T.; Nilsson, A.; Allen, J.F. Photosynthetic control of chloroplast gene expression. Nature 1999, 397, 625–628. [Google Scholar]
- Moseley, J.L.; Allinger, T.; Herzog, S.; Hoerth, P.; Wehinger, E.; Merchant, S.; Hippler, M. Adaptation to Fe-deficiency requires remodeling of the photosynthetic apparatus. EMBO J. 2002, 21, 6709–6720. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Leister, D.; Bolle, C. Photosynthetic lesions can trigger accelerated senescence in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 6891–6903. [Google Scholar] [CrossRef] [PubMed]
- Chi, W.; Sun, X.; Zhang, L. Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 2013, 64, 559–582. [Google Scholar] [CrossRef] [PubMed]
- Larkin, R.M.; Ruckle, M.E. Integration of light and plastid signals. Curr. Opin. Plant Biol. 2008, 11, 593–599. [Google Scholar] [CrossRef] [PubMed]
- Gollan, P.J.; Tikkanen, M.; Aro, E.M. Photosynthetic light reactions: Integral to chloroplast retrograde signalling. Curr. Opin. Plant Biol. 2015, 27, 180–191. [Google Scholar] [CrossRef] [PubMed]
- De Barajas-López, J.D.; Blanco, N.E.; Strand, Å. Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim. Biophys. Acta 2013, 1833, 425–437. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Karpinska, B.; Krupinska, K. The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: A hypothesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2014, 369, 20130226. [Google Scholar] [CrossRef] [PubMed]
- Pfannschmidt, T. Plastidial retrograde signalling—A true “plastid factor” or just metabolite signatures? Trends Plant Sci. 2010, 15, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, E.; Miao, Y.; Mulisch, M.; Krupinska, K. Single-stranded DNA-binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 2008, 147, 1800–1804. [Google Scholar] [CrossRef] [PubMed]
- Krupinska, K.; Haussuhl, K.; Schafer, A.; van der Kooij, T.A.; Leckband, G.; Lorz, H.; Falk, J. A novel nucleus-targeted protein is expressed in barley leaves during senescence and pathogen infection. Plant Physiol. 2002, 130, 1172–1180. [Google Scholar] [CrossRef] [PubMed]
- Desveaux, D.; Maréchal, A.; Brisson, N. Whirly transcription factors: Defense gene regulation and beyond. Trends Plant Sci. 2005, 10, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Pfalz, J.; Liere, K.; Kandlbinder, A.; Dietz, K.J.; Oelmuller, R. pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 2006, 18, 176–197. [Google Scholar] [CrossRef] [PubMed]
- Marechal, A.; Parent, J.S.; Veronneau-Lafortune, F.; Joyeux, A.; Lang, B.F.; Brisson, N. Whirly proteins maintain plastid genome stability in Arabidopsis. Proc. Natl. Acad. Sci. USA 2009, 106, 14693–14698. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Marechal, A.; Parent, J.S.; Lepage, E.; Sygusch, J.; Brisson, N. Crystal structures of DNA-Whirly complexes and their role in Arabidopsis organelle genome repair. Plant Cell 2010, 22, 1849–1867. [Google Scholar] [CrossRef] [PubMed]
- Lepage, E.; Zampini, E.; Brisson, N. Plastid genome instability leads to reactive oxygen species production and plastid-to-nucleus retrograde signaling in Arabidopsis. Plant Physiol. 2013, 163, 867–881. [Google Scholar] [CrossRef] [PubMed]
- Kucharewicz, W.; Distelfeld, A.; Bilger, W.; Muller, M.; Munne-Bosch, S.; Hensel, G.; Krupinska, K. Acceleration of leaf senescence is slowed down in transgenic barley plants deficient in the DNA/RNA-binding protein WHIRLY1. J. Exp. Bot. 2017, 68, 983–996. [Google Scholar] [CrossRef] [PubMed]
- Ruckle, M.E.; Burgoon, L.D.; Lawrence, L.A.; Sinkler, C.A.; Larkin, R.M. Plastids are major regulators of light signaling in Arabidopsis. Plant Physiol. 2012, 159, 366–390. [Google Scholar] [CrossRef] [PubMed]
- Comadira, G.; Rasool, B.; Kaprinska, B.; Garcia, B.M. WHIRLY1 Functions in the Control of Responses to Nitrogen Deficiency but Not Aphid Infestation in Barley. Plant Physiol. 2015, 168, 1140–1151. [Google Scholar] [CrossRef] [PubMed]
- Gregersen, P.L.; Foyer, C.H.; Krupinska, K. Photosynthesis and Leaf Senescence as Determinants of Plant Productivity. In Biotechnological Approaches to Barley Improvement; Kumlehn, J., Stein, N., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 113–138. [Google Scholar]
- Miao, Y.; Jiang, J.; Ren, Y.; Zhao, Z. The single-stranded DNA binding protein WHIRLY1 represses WRKY53 expression and delays leaf senescence in a developmental stage-dependent manner in Arabidopsis thaliana. Plant Physiol. 2013, 163, 746–756. [Google Scholar] [CrossRef] [PubMed]
- Tikkanen, M.; Rantala, S.; Grieco, M.; Aro, E.M. Comparative analysis of mutant plants impaired in the main regulatory mechanisms of photosynthetic light reactions—From biophysical measurements to molecular mechanisms. Plant Physiol. Biochem. 2017, 112, 290–301. [Google Scholar] [CrossRef] [PubMed]
- Benson, S.L.; Maheswaran, P.; Ware, M.A.; Hunter, C.N.; Horton, P.; Jansson, S.; Ruban, A.V.; Johnson, M.P. An intact light harvesting complex I antenna system is required for complete state transitions in Arabidopsis. Nat. Plants 2015, 1, 15176. [Google Scholar] [CrossRef] [PubMed]
- Tongra, T.; Bharti, S.; Jajoo, A. Cyclic electron flow around photosystem I is enhanced at low pH. Plant Physiol. Biochem. 2014, 83, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Croce, R.; Morosinotto, T.; Castelletti, S.; Breton, J.; Bassi, R. The Lhca antenna complexes of higher plants photosystem I. Biochim. Biophys. Acta 2002, 1556, 29–40. [Google Scholar] [CrossRef]
- Wientjes, E.; Oostergetel, G.T.; Jansson, S.; Boekema, E.J.; Croce, R. The role of Lhca complexes in the supramolecular organization of higher plant photosystem I. J. Biol. Chem. 2009, 284, 7803–7810. [Google Scholar] [CrossRef] [PubMed]
- Busch, A.; Hippler, M. The structure and function of eukaryotic photosystem I. Biochim. Biophys. Acta 2011, 1807, 864–877. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Li, Y.; Jiang, Y.; Wu, B.; Miao, Y. Phosphorylation of WHIRLY1 by CIPK14 Shifts Its Localization and Dual Functions in Arabidopsis. Mol. Plant 2017, 10, 749–763. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Drory, O.; Nelson, N. The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 2007, 447, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.E.; Bassi, R.; Boekema, E.J.; Dekker, J.P.; Jansson, S.; Leister, D.; Robinson, C.; Scheller, H.V. Structure, function and regulation of plant photosystem I. Biochim. Biophys. Acta 2007, 1767, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Kouril, R.; Strouhal, O.; Nosek, L.; Lenobel, R.; Chamrad, I.; Boekema, E.J.; Sebela, M.; Ilik, P. Structural characterization of a plant photosystem I and NAD(P)H dehydrogenase supercomplex. Plant J. 2014, 77, 568–576. [Google Scholar] [CrossRef] [PubMed]
- Rumeau, D.; Becuwe-Linka, N.; Beyly, A.; Louwagie, M.; Garin, J.; Peltier, G. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid NDH complex functioning in higher plants. Plant Cell 2005, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Fukao, Y.; Fujiwara, M.; Takami, T.; Shikanai, T. Efficient operation of NAD(P)H dehydrogenase requires supercomplex formation with photosystem I via minor LHCI in Arabidopsis. Plant Cell 2009, 21, 3623–3640. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Shikanai, T. Supercomplex formation with photosystem I is required for the stabilization of the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Physiol. 2011, 155, 1629–1639. [Google Scholar] [CrossRef] [PubMed]
- Roach, T.; Krieger-Liszkay, A. Regulation of photosynthetic electron transport and photoinhibition. Curr. Protein Pept. Sci. 2014, 15, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Chuang-Dao, J.; Hul-Yuan, G.; Qi, Z.; Lei, S. Effects of Iron Deficiency on Photosynthesis and Photosystem II Function in Soybean Leaf. J. Plant Physiol. Mol. Biol. 2007, 1, 53–60. [Google Scholar]
- Scheller, H.V.; Jensen, P.E.; Haldrup, A.; Lunde, C.; Knoetzel, J. Role of subunits in eukaryotic Photosystem I. Biochim. Biophys. Acta 2001, 1507, 41–60. [Google Scholar] [CrossRef]
- Thomas, D.J.; Thomas, J.; Youderian, P.A.; Herbert, S.K. Photoinhibition and light-induced cyclic electron transport in ndhB- and psaE- mutants of Synechocystis sp. PCC 6803. Plant Cell Physiol. 2001, 42, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Rossel, J.B.; Wilson, I.W.; Pogson, B.J. Global changes in gene expression in response to high light in Arabidopsis. Plant Physiol. 2002, 130, 1109–1120. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Jiang, Y.; Yu, D. WRKY22 transcription factor mediates dark-induced leaf senescence in Arabidopsis. Mol. Cells 2011, 31, 303–313. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Park, J.H.; Lee, M.H.; Yu, J.H.; Kim, S.Y. Isolation and functional characterization of CE1 binding proteins. BMC Plant Biol. 2010, 10, 277. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Perez, S.; Gutierrez, J.; Garcia-Garcia, F.; Osuna, D.; Dopazo, J.; Lorenzo, O.; Revuelta, J.L.; Arellano, J.B. Early transcriptional defense responses in Arabidopsis cell suspension culture under high-light conditions. Plant Physiol. 2011, 156, 1439–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Cao, G.; Wang, X.; Miao, J.; Liu, X.; Chen, Z.; Qu, L.J.; Gu, H. Identification and characterization of COI1-dependent transcription factor genes involved in JA-mediated response to wounding in Arabidopsis plants. Plant Cell Rep. 2008, 27, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Matsuo, M.; Oelmuller, R. REDOX RESPONSIVE TRANSCRIPTION FACTOR1 is involved in age-dependent and systemic stress signaling. Plant Signal. Behav. 2015, 10, e1051279. [Google Scholar] [CrossRef] [PubMed]
- Passardi, F.; Tognolli, M.; De Meyer, M.; Penel, C.; Dunand, C. Two cell wall associated peroxidases from Arabidopsis influence root elongation. Planta 2006, 223, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.H.; Kim, H.J.; Ryu, J.S.; Choi, H.; Jeong, S.; Shin, J.; Choi, G.; Nam, H.G. CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. Plant J. 2008, 55, 361–371. [Google Scholar] [CrossRef] [PubMed]
- Cappadocia, L.; Parent, J.S.; Sygusch, J.; Brisson, N. A family portrait: Structural comparison of the Whirly proteins from Arabidopsis thaliana and Solanum tuberosum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 1207–1211. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Takusagawa, M.; Harada, N.; Fukao, Y.; Yamaoka, S.; Kohchi, T.; Hori, K.; Ohta, H.; Shikanai, T.; Nishimura, Y. Eukaryotic Components Remodeled Chloroplast Nucleoid Organization during the Green Plant Evolution. Genome Biol. Evol. 2015, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Bertamini, M.; Muthuchelian, K.; Nedunchezhian, N. Iron deficiency induced changes on the donor side of PS II in field grown grapevine (Vitis vinifera L. cv. Pinot noir) leaves. Plant Sci. 2002, 162, 599–605. [Google Scholar] [CrossRef]
- Cui, Y.; Tian, Z.; Zhang, X.; Muhammad, A.; Han, H.; Jiang, D.; Cao, W.; Dai, T. Effect of water deficit during vegetative growth periods on post-anthesis photosynthetic capacity and grain yield in winter wheat (Triticum aestivum L.). Acta Physiol. Plant. 2015, 37, 196. [Google Scholar] [CrossRef]
- Ihalainen, J.A.; Klimmek, F.; Ganeteg, U.; van Stokkum, I.H.; van Grondelle, R.; Jansson, S.; Dekker, J.P. Excitation energy trapping in photosystem I complexes depleted in Lhca1 and Lhca4. FEBS Lett. 2005, 579, 4787–4791. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.G.; Hwang, S.G.; Park, Y.C.; Park, H.M.; Kim, D.S.; Park, D.H.; Jang, C.S. Molecular characterization of the cold- and heat-induced Arabidopsis PXL1 gene and its potential role in transduction pathways under temperature fluctuations. J. Plant Physiol. 2015, 176, 138–146. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.E.; Rosgaard, L.; Knoetzel, J.; Scheller, H.V. Photosystem I activity is increased in the absence of the PSI-G subunit. J. Biol. Chem. 2002, 277, 2798–2803. [Google Scholar] [CrossRef] [PubMed]
- Alonso, J.M.; Stepanova, A.N.; Leisse, T.J.; Kim, C.J.; Chen, H.; Shinn, P.; Stevenson, D.K.; Zimmerman, J.; Barajas, P.; Cheuk, R.; et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 2003, 301, 653–657. [Google Scholar] [CrossRef] [PubMed]
- Klughammer, C.; Schreiber, U. Saturation pulse method for assessment of energy conversion in PS I. PAM Appl. Notes 2008, 1, 11–14. [Google Scholar]
- Maxwell, K.; Johnson, G.N. Chlorophyll fluorescence—A practical guide. J. Exp. Bot. 2000, 51, 659–668. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Ohmori, Y.; Ratel, E. High root temperature blocks both linear and cyclic electron transport in the dark during chilling of the leaves of rice seedlings. Plant Cell Physiol. 2011, 52, 1697–1707. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Chen, F.; Mu, S.; Zhang, D.; Pan, X.; Lee, D.J. Simultaneous analysis of photosystem responses of Microcystis aeruginoga under chromium stress. Ecotoxicol. Environ. Saf. 2013, 88, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Shao, L.; Shu, Z.; Peng, C.-L.; Lin, Z.-F.; Yang, C.-W.; Gu, Q. Enhanced sensitivity of Arabidopsis anthocyanin mutants to photooxidation: A study with fluorescence imaging. Funct. Plant Biol. 2008, 35, 714–724. [Google Scholar] [CrossRef]
- Miao, Y.; Zentgraf, U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell 2007, 19, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Miao, Y.; Zentgraf, U. A HECT E3 ubiquitin ligase negatively regulates Arabidopsis leaf senescence through degradation of the transcription factor WRKY53. Plant J. 2010, 63, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Fling, S.P.; Gregerson, D.S. Peptide and protein molecular weight determination by electrophoresis using a high-molarity tris buffer system without urea. Anal. Biochem. 1986, 155, 83–88. [Google Scholar] [CrossRef]
- Poulsen, C. The barley chloroplast genome: Physical structure and transcriptional activity in vivo. Carlsberg Res. Commun. 1983, 48, 57–80. [Google Scholar] [CrossRef]
- Caffarri, S.; Croce, R.; Breton, J.; Bassi, R. The major antenna complex of photosystem II has a xanthophyll binding site not involved in light harvesting. J. Biol. Chem. 2001, 276, 35924–35933. [Google Scholar] [CrossRef] [PubMed]
- Eshaghi, S.; Andersson, B.; Barber, J. Isolation of a highly active PSII-LHCII supercomplex from thylakoid membranes by a direct method. FEBS Lett. 1999, 446, 23–26. [Google Scholar] [CrossRef]
- Bassi, R.; Simpson, D. Chlorophyll-protein complexes of barley photosystem I. Eur. J. Biochem. 1987, 163, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Gruissem, W.; Greenberg, B.M.; Zurawski, G.; Hallick, R.B. Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 1986, 118, 253–270. [Google Scholar] [PubMed]
- Krause, K.; Kilbienski, I.; Mulisch, M.; Rödiger, A.; Schäfer, A.; Krupinska, K. DNA-binding proteins of the Whirly family in Arabidopsis thaliana are targeted to the organelles. FEBS Lett. 2005, 579, 3707–3712. [Google Scholar] [CrossRef] [PubMed]
- Bilger, W.; Heber, U.; Schreiber, U. Kinetic Relationship between Energy-Dependent Fluorescence Quenching, Light Scattering, Chlorophyll Luminescence and Proton Pumping in Intact Leaves. Z. Naturforsch. 1988, 43, 877–887. [Google Scholar]
- Peng, L.; Fukao, Y.; Fujiwara, M.; Shikanai, T. Multistep assembly of chloroplast NADH dehydrogenase-like subcomplex A requires several nucleus-encoded proteins, including CRR41 and CRR42, in Arabidopsis. Plant Cell 2012, 24, 202–214. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T.; Endo, T.; Hashimoto, T.; Yamada, Y.; Asada, K.; Yokota, A. Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 1998, 95, 9705–9709. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Pandeya, D.; Nath, K.; Zulfugarov, I.S.; Yoo, S.C.; Zhang, H.; Yoo, J.H.; Cho, S.H.; Koh, H.J.; Kim, D.S.; et al. ZEBRA-NECROSIS, a thylakoid-bound protein, is critical for the photoprotection of developing chloroplasts during early leaf development. Plant J. 2010, 62, 713–725. [Google Scholar] [CrossRef] [PubMed]








© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, D.; Lin, W.; Deng, B.; Ren, Y.; Miao, Y. Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis. Int. J. Mol. Sci. 2017, 18, 2352. https://doi.org/10.3390/ijms18112352
Huang D, Lin W, Deng B, Ren Y, Miao Y. Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis. International Journal of Molecular Sciences. 2017; 18(11):2352. https://doi.org/10.3390/ijms18112352
Chicago/Turabian StyleHuang, Dongmei, Wenfang Lin, Ban Deng, Yujun Ren, and Ying Miao. 2017. "Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis" International Journal of Molecular Sciences 18, no. 11: 2352. https://doi.org/10.3390/ijms18112352
APA StyleHuang, D., Lin, W., Deng, B., Ren, Y., & Miao, Y. (2017). Dual-Located WHIRLY1 Interacting with LHCA1 Alters Photochemical Activities of Photosystem I and Is Involved in Light Adaptation in Arabidopsis. International Journal of Molecular Sciences, 18(11), 2352. https://doi.org/10.3390/ijms18112352