What Has Been Seen Cannot Be Unseen—Detecting Auxin In Vivo
Abstract
:1. Introduction
2. Indirect Auxin Visualization—Methods Based on Detection of Auxin Action
2.1. Reporters Based on Auxin Signalling
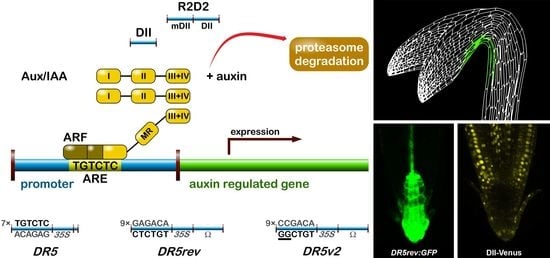
2.1.1. The Signalling Reporter DR5 and Variants
2.1.2. Degradation-Based Auxin Reporters
2.1.3. Dissecting the Specificity of the Auxin Signalling
2.2. Focused on Auxin Source
2.3. Following Auxin Flow
2.4. Immunolocalisation and In Situ Hybridisation Approaches
3. Direct Methods for Tracking Auxin Distribution
3.1. Immunolocalisation of IAA
3.2. Radiolabelling
3.2.1. Traditional Methods for Studying Polar Auxin Transport in Plants
3.2.2. Cellular Polar Auxin Transport Matters
3.3. Fluorescent Labelling
3.3.1. Strategies to Label Plant Hormones
3.3.2. Up-To-Date Labelling of Auxins
3.4. Microelectrodes
4. New Valuable Tools to Visualize Auxin Metabolites
4.1. Cell-Type Specific Mass Spectrometric Analysis
4.2. Auxin Monitoring by Solid-State Biosensors
5. Future Prospects
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Davies, P.J. Plant Hormones—Biosynthesis, Signal Transduction, Action! 3rd ed.; Springer: London, UK, 2010; pp. 16–35. ISBN 9788578110796. [Google Scholar]
- Korasick, D.A.; Westfall, C.S.; Lee, S.G.; Nanao, M.H.; Dumas, R.; Hagen, G.; Guilfoyle, T.J.; Jez, J.M.; Strader, L.C. Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression. Proc. Natl. Acad. Sci. USA 2014, 111, 5427–5432. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Estelle, M. Diversity and specificity: Auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 2014, 21, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Powers, S.K.; Strader, L.C. Up in the air: Untethered Factors of Auxin Response. F1000Research 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Löbler, M.; Klämbt, D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). I. Purification by immunological methods and characterization. J. Biol. Chem. 1985, 260, 9848–9853. [Google Scholar] [PubMed]
- Jones, A.M.; Venis, M.A. Photoaffinity labeling of indole-3-acetic acid-binding proteins in maize. Proc. Natl. Acad. Sci. USA 1989, 86, 6153–6156. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zhang, Y.; Zhang, D.; Dai, X.; Estelle, M.; Zhao, Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc. Natl. Acad. Sci. USA 2015, 112, 2275–2280. [Google Scholar] [CrossRef] [PubMed]
- Paque, S.; Weijers, D. Q&A: Auxin: The plant molecule that influences almost anything. BMC Biol. 2016, 14, 67. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K. Auxin metabolism and homeostasis during plant development. Development 2013, 140, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Pěnčík, A.; Simonovik, B.; Petersson, S.V.; Henyková, E.; Simon, S.; Greenham, K.; Zhang, Y.; Kowalczyk, M.; Estelle, M.; Zažímalová, E.; et al. Regulation of auxin homeostasis and gradients in Arabidopsis roots through the formation of the indole-3-acetic acid catabolite 2-oxindole-3-acetic acid. Plant Cell 2013, 25, 3858–3870. [Google Scholar] [CrossRef] [PubMed]
- Normanly, J. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harb. Perspect. Biol. 2010, 2, a001594. [Google Scholar] [CrossRef] [PubMed]
- Ludwig-Müller, J. Auxin conjugates: Their role for plant development and in the evolution of land plants. J. Exp. Bot. 2011, 62, 1757–1773. [Google Scholar] [CrossRef] [PubMed]
- Reemmer, J.; Murphy, A.S. Intercellular Transport of Auxin. In Auxin and Its Role in Plant Development, 1st ed.; Zažímalová, E., Petrášek, J., Benková, E., Eds.; Springer: London, UK, 2014; Volume 33, pp. 75–101. ISBN 978-3-7091-1526-8. [Google Scholar]
- Swarup, R.; Péret, B. AUX/LAX family of auxin influx carriers—An overview. Front. Plant Sci. 2012, 3, 225. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Friml, J. PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Do, T.H.T.; Martinoia, E.; Lee, Y. Functions of ABC transporters in plant growth and development. Curr. Opin. Plant Biol. 2018, 41, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Barbez, E.; Kubeš, M.; Rolčík, J.; Béziat, C.; Pěnčík, A.; Wang, B.; Rosquete, M.R.; Zhu, J.; Dobrev, P.I.; Lee, Y.; et al. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature 2012, 485, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Vieten, A.; Sauer, M.; Brewer, P.B.; Friml, J. Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci. 2007, 12, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; Friml, J. The march of the PINs: Developmental plasticity by dynamic polar targeting in plant cells. EMBO J. 2010, 29, 2700–2714. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, J.; Friml, J. Auxin transport routes in plant development. Development 2009, 136, 2675–2688. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.J.; Marchant, A.; Green, H.G.; May, S.T.; Ward, S.P.; Millner, P.A.; Walker, A.R.; Schulz, B.; Feldmann, K.A. Arabidopsis AUX1 Gene: A Permease-Like Regulator of Root Gravitropism. Science 1996, 273, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Stone, B.B.; Stowe-Evans, E.L.; Harper, R.M.; Brandon Celaya, R.; Ljung, K.; Sandberg, G.; Liscum, E. Disruptions in AUX1-dependent auxin influx alter hypocotyl phototropism in Arabidopsis. Mol. Plant 2008, 1, 129–144. [Google Scholar] [CrossRef] [PubMed]
- Swarup, K.; Benková, E.; Swarup, R.; Casimiro, I.; Péret, B.; Yang, Y.; Parry, G.; Nielsen, E.; De Smet, I.; Vanneste, S.; et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat. Cell Biol. 2008, 10, 946–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, A.R.; Kramer, E.M.; Knox, K.; Swarup, R.; Bennett, M.J.; Lazarus, C.M.; Leyser, H.M.O.; Grierson, C.S. Auxin transport through non-hair cells sustains root-hair development. Nat. Cell Biol. 2009, 11, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Peer, W.A.; Murphy, A.S. Flavonoids and auxin transport: Modulators or regulators? Trends Plant Sci. 2007, 12, 556–563. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, W.; Klíma, P.; Quareshy, M.; Cesarino, I.; Kumpf, R.P.; Corneillie, S.; Araújo, P.; Viaene, T.; Goeminne, G.; Nowack, M.K.; et al. cis-Cinnamic Acid Is a Novel, Natural Auxin Efflux Inhibitor that Promotes Lateral Root Formation. Plant Physiol. 2017, 173, 552–565. [Google Scholar] [CrossRef] [PubMed]
- Brown, D.E.; Rashotte, A.M.; Murphy, A.S.; Normanly, J.; Tague, B.W.; Peer, W.A.; Taiz, L.; Mudaye, G.K. Flavonoids act as negative regulators of auxin transport in vivo in arabidopsis. Plant Physiol. 2001, 126, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, B.M.; Geisler, M.; Bigler, L.; Ringli, C. Flavonols Accumulate Asymmetrically and Affect Auxin Transport in Arabidopsis. Plant Physiol. 2011, 156, 585–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santelia, D.; Henrichs, S.; Vincenzetti, V.; Sauer, M.; Bigler, L.; Klein, M.; Bailly, A.; Lee, Y.; Friml, J.; Geisler, M.; et al. Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J. Biol. Chem. 2008, 283, 31218–31226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peer, W.A.; Bandyopadhyay, A.; Blakeslee, J.J.; Makam, S.N.; Chen, R.J.; Masson, P.H.; Murphy, A.S. Variation in Expression and Protein Localization of the PIN Family of Auxin Efflux Facilitator Proteins in Flavonoid Mutants with Altered Auxin Transport in Arabidopsis thaliana. Plant Cell 2004, 16, 1898–1911. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Blakeslee, J.J.; Bouchard, R.; Lee, O.R.; Vincenzetti, V.; Bandyopadhyay, A.; Titapiwatanakun, B.; Peer, W.A.; Bailly, A.; Richards, E.L.; et al. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005, 44, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.J.; Bandyopadhyay, A.; Lee, O.R.; Mravec, J.; Titapiwatanakun, B.; Sauer, M.; Makam, S.N.; Cheng, Y.; Bouchard, R.; Adamec, J.; et al. Interactions among PIN-FORMED and P-glycoprotein auxin transporters in Arabidopsis. Plant Cell 2007, 19, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Grunewald, W.; De Smet, I.; Lewis, D.R.; Löfke, C.; Jansen, L.; Goeminne, G.; Bosschea, R.V.; Karimi, M.; De Rybela, B.; Vanholmea, B.; et al. Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 109, 1554–1559. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Ramirez, M.V.; Miller, N.D.; Vallabhaneni, P.; Ray, W.K.; Helm, R.F.; Winkel, B.S.J.; Muday, G.K. Auxin and Ethylene Induce Flavonol Accumulation through Distinct Transcriptional Networks. Plant Physiol. 2011, 156, 144–164. [Google Scholar] [CrossRef] [PubMed]
- Titapiwatanakun, B.; Murphy, A.S. Post-transcriptional regulation of auxin transport proteins: Cellular trafficking, protein phosphorylation, protein maturation, ubiquitination, and membrane composition. J. Exp. Bot. 2009, 60, 1093–1107. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hagen, G.; Guilfoyle, T. An Auxin-Responsive Promoter Is Differentially Induced by Auxin Gradients during Tropisms. Plant Cell 1991, 3, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Larkin, P.J.; Gibson, J.M.; Mathesius, U.; Weinman, J.J.; Gartner, E.; Hall, E.; Tanner, G.J.; Rolfe, B.G.; Djordjevic, M.A. Transgenic white clover. Studies with the auxin-responsive promoter, GH3, in root gravitropism and lateral root development. Transgenic Res. 1996, 5, 443–450. [Google Scholar] [CrossRef]
- Ballas, N.; Wong, L.M.; Theologist, A. Identification of the Auxin-responsive Element, AuxRE, in the Primary indoleacetic Acid-inducible Gene, PS-IAA4/5, of Pea (Pisum sativum). J. Mol. Biol. 1993, 233, 580–596. [Google Scholar] [CrossRef] [PubMed]
- Ballas, N.; Wong, L.M.; Ke, M.; Theologis, A. Two auxin-responsive domains interact positively to induce expression of the early indoleacetic acid-inducible gene PS-IAA4/5. Proc. Natl. Acad. Sci. USA 1995, 92, 3483–3487. [Google Scholar] [CrossRef] [PubMed]
- Oono, Y.; Chen, Q.G.; Overvoorde, P.J.; Köhler, C.; Theologis, A. age Mutants of Arabidopsis exhibit altered auxin-regulated gene expression. Plant Cell 1998, 10, 1649–1662. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, J.I.; Yuan, S.; Dale, J.M.; Tanner, V.N.; Theologis, A. Identification of inhibitors of auxin transcriptional activation by means of chemical genetics in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 14978–14983. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Murfett, J.; Hagen, G.; Guilfoyle, T.J. Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 1997, 9, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Vieten, A.; Sauer, M.; Weijers, D.; Schwarz, H.; Hamann, T.; Offringa, R.; Jürgens, G. Efflux-dependent auxin gradients establish the apical–basal axis of Arabidopsis. Nature 2003, 426, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Martin, G.; Li, Y.; Guilfoyle, T.J. Auxin-induced expression of the soybean GH3 promoter in transgenic tobacco plants. Plant Mol. Biol. 1991, 17, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Hagen, G.; Guilfoyle, T.J. Rapid induction of selective transcription by auxins. Mol. Cell. Biol. 1985, 5, 1197–1203. [Google Scholar] [CrossRef] [PubMed]
- Ulmasov, T.; Liu, Z.B.; Hagen, G.; Guilfoyle, T.J. Composite structure of auxin response elements. Plant Cell 1995, 7, 1611–1623. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.B.; Ulmasov, T.; Shi, X.; Hagen, G.; Guilfoyle, T.J. Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 1994, 6, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Z.B.; Shi, X.; Hagen, G.; Guilfoyle, T.J. An auxin-inducible element in soybean SAUR promoters. Plant Physiol. 1994, 106, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Friml, J.; Benková, E.; Blilou, I.; Wisniewska, J.; Hamann, T.; Ljung, K.; Woody, S.; Sandberg, G.; Scheres, B.; Jürgens, G.; et al. AtPIN4 mediates sink driven auxin gradients and patterning in Arabidopsis roots. Cell 2002, 108, 661–673. [Google Scholar] [CrossRef]
- Heisler, M.G.; Ohno, C.; Das, P.; Sieber, P.; Reddy, G.V.; Long, J.A.; Meyerowitz, E.M. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 2005, 15, 1899–1911. [Google Scholar] [CrossRef] [PubMed]
- Gallavotti, A.; Yang, Y.; Schmidt, R.J.; Jackson, D. The Relationship between Auxin Transport and Maize Branching. Plant Physiol. 2008, 147, 1913–1923. [Google Scholar] [CrossRef] [PubMed]
- Marin, E.; Jouannet, V.; Herz, A.; Lokerse, A.S.; Weijers, D.; Vaucheret, H.; Nussaume, L.; Crespi, M.D.; Maizel, A. miR390, Arabidopsis TAS3 tasiRNAs, and Their AUXIN RESPONSE FACTOR Targets Define an Autoregulatory Network Quantitatively Regulating Lateral Root Growth. Plant Cell 2010, 22, 1104–1117. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Van Norman, J.M.; Moreno, A.; Zhang, J.; Ahnert, S.E.; Benfey, P.N. Oscillating Gene Expression Determines Competence for Periodic Arabidopsis Root Branching. Science 2010, 329, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Benková, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertová, D.; Jürgens, G.; Friml, J. Local, Efflux-Dependent Auxin Gradients as a Common Module for Plant Organ Formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Boer, D.R.; Freire-Rios, A.; van den Berg, W.A.M.; Saaki, T.; Manfield, I.W.; Kepinski, S.; López-Vidrieo, I.; Franco-Zorrilla, J.M.; de Vries, S.C.; Solano, R.; et al. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 2014, 156, 577–589. [Google Scholar] [CrossRef] [PubMed]
- Korber, H.; Strizhov, N.; Staiger, D.; Feldwisch, J.; Olsson, O.; Sandberg, G.; Palme, K.; Schell, J.; Koncz, C. T-DNA gene 5 of Agrobacterium modulates auxin response by autoregulated synthesis of a growth hormone antagonist in plants. EMBO J. 1991, 10, 3983–3991. [Google Scholar] [PubMed]
- Liao, C.Y.; Smet, W.; Brunoud, G.; Yoshida, S.; Vernoux, T.; Weijers, D. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods 2015, 12, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Scarpella, E.; Marcos, D.; Friml, J.; Berleth, T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006, 20, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef] [PubMed]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; van den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, X.; Calderon-Villalobos, L.I.A.; Sharon, M.; Zheng, C.; Robinson, C.V.; Estelle, M.; Zheng, N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar] [CrossRef] [PubMed]
- Brunoud, G.; Wells, D.M.; Oliva, M.; Larrieu, A.; Mirabet, V.; Burrow, A.H.; Beeckman, T.; Kepinski, S.; Traas, J.; Bennett, M.J.; et al. A novel sensor to map auxin response and distribution at high spatio-temporal resolution. Nature 2012, 482, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.; Middleton, A.M.; French, A.P.; Brunoud, G.; Sato, E.M.; Wilson, M.H.; Peret, B.; et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 4668–4673. [Google Scholar] [CrossRef] [PubMed]
- Pernisova, M.; Prat, T.; Grones, P.; Harustiakova, D.; Matonohova, M.; Spichal, L.; Nodzynski, T.; Friml, J.; Hejatko, J. Cytokinins influence root gravitropism via differential regulation of auxin transporter expression and localization in Arabidopsis. New Phytol. 2016, 212, 497–509. [Google Scholar] [CrossRef] [PubMed]
- Von Wangenheim, D.; Hauschild, R.; Friml, J. Light Sheet Fluorescence Microscopy of Plant Roots Growing on the Surface of a Gel. J. Vis. Exp. 2017. [Google Scholar] [CrossRef] [PubMed]
- Wend, S.; Bosco, C.D.; Kämpf, M.M.; Ren, F.; Palme, K.; Weber, W.; Dovzhenko, A.; Zurbriggen, M.D. A quantitative ratiometric sensor for time-resolved analysis of auxin dynamics. Sci. Rep. 2013, 3, 2052. [Google Scholar] [CrossRef] [PubMed]
- Rademacher, E.H.; Möller, B.; Lokerse, A.S.; Llavata-Peris, C.I.; van Den Berg, W.; Weijers, D. A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 2011, 68, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Abel, S.; Oeller, P.W.; Theologist, A. Early auxin-induced genes encode short-lived nuclear proteins. Biochemistry 1994, 91, 326–330. [Google Scholar] [CrossRef]
- Gray, W.M.; Kepinski, S.; Rouse, D.; Leyser, O.; Estelle, M. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 2001, 414, 271–276. [Google Scholar] [CrossRef] [PubMed]
- Swarup, R.; Friml, J.; Marchant, A.; Ljung, K.; Sandberg, G.; Palme, K.; Bennett, M. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 2001, 15, 2648–2653. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Uhlir, N.J.; Reed, J.W. Arabidopsis SHY2/IAA3 Inhibits Auxin-Regulated Gene Expression. Plant Cell 2002, 14, 301–319. [Google Scholar] [CrossRef] [PubMed]
- Weijers, D.; Benkova, E.; Jäger, K.E.; Schlereth, A.; Hamann, T.; Kientz, M.; Wilmoth, J.C.; Reed, J.W.; Jürgens, G. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J. 2005, 24, 1874–1885. [Google Scholar] [CrossRef] [PubMed]
- Ploense, S.E.; Wu, M.F.; Nagpal, P.; Reed, J.W. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 2009, 136, 1509–1517. [Google Scholar] [CrossRef] [PubMed]
- De Rybel, B.; Vassileva, V.; Parizot, B.; Demeulenaere, M.; Grunewald, W.; Audenaert, D.; Van Campenhout, J.; Overvoorde, P.; Jansen, L.; Vanneste, S.; et al. A novel Aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Curr. Biol. 2010, 20, 1697–1706. [Google Scholar] [CrossRef] [PubMed]
- Müller, C.J.; Valdés, A.E.; Wang, G.; Ramachandran, P.; Beste, L.; Uddenberg, D.; Carlsbecker, A. PHABULOSA Mediates an Auxin Signaling Loop to Regulate Vascular Patterning in Arabidopsis. Plant Physiol. 2016, 170, 956–970. [Google Scholar] [CrossRef] [PubMed]
- Winkler, M.; Niemeyer, M.; Hellmuth, A.; Janitza, P.; Christ, G.; Samodelov, S.L.; Wilde, V.; Majovsky, P.; Trujillo, M.; Zurbriggen, M.D.; et al. Variation in auxin sensing guides AUX/IAA transcriptional repressor ubiquitylation and destruction. Nat. Commun. 2017, 8, 15706. [Google Scholar] [CrossRef] [PubMed]
- Piya, S.; Shrestha, S.K.; Binder, B.; Stewart, C.N.; Hewezi, T. Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front. Plant Sci. 2014, 5, 744. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, H. Current aspects of auxin biosynthesis in plants. Biosci. Biotechnol. Biochem. 2016, 80, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Stepanova, A.N.; Robertson-Hoyt, J.; Yun, J.; Benavente, L.M.; Xie, D.Y.; Doležal, K.; Schlereth, A.; Jürgens, G.; Alonso, J.M. TAA1-Mediated Auxin Biosynthesis Is Essential for Hormone Crosstalk and Plant Development. Cell 2008, 133, 177–191. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin Synthesized by the YUCCA Flavin Monooxygenases Is Essential for Embryogenesis and Leaf Formation in Arabidopsis. Plant Cell 2007, 19, 2430–2439. [Google Scholar] [CrossRef] [PubMed]
- Robert, H.S.; Grones, P.; Stepanova, A.N.; Robles, L.M.; Lokerse, A.S.; Alonso, J.M.; Weijers, D.; Friml, J. Local auxin sources orient the apical-basal axis in Arabidopsis embryos. Curr. Biol. 2013, 23, 2506–2512. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; de-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin Overproduction in Shoots Cannot Rescue Auxin Deficiencies in Arabidopsis Roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertová, D.; Wisniewska, J.; Tadele, Z.; Kubeš, M.; Covanová, M.; et al. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Zažímalová, E.; Murphy, A.S.; Yang, H.; Hoyerova, K.; Hošek, P. Auxin Transporters—Why So Many? Cold Spring Harb. Perspect. Biol. 2010, 2, a001552. [Google Scholar] [CrossRef] [PubMed]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Vieten, A.; Vanneste, S.; Wis, J.; Benková, E.; Benjamins, R.; Beeckman, T.; Luschnig, C. Functional redundancy of PIN proteins is accompanied by auxin-dependent cross-regulation of PIN expression. Development 2005, 132, 4521–4531. [Google Scholar] [CrossRef] [PubMed]
- Xu, J. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 Function in Epidermal Cell Polarity. Plant Cell 2005, 17, 525–536. [Google Scholar] [CrossRef] [PubMed]
- Zadnikova, P.; Petrasek, J.; Marhavy, P.; Raz, V.; Vandenbussche, F.; Ding, Z.; Schwarzerova, K.; Morita, M.T.; Tasaka, M.; Hejatko, J.; et al. Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 2010, 137, 607–617. [Google Scholar] [CrossRef] [PubMed]
- Mravec, J.; Skůpa, P.; Bailly, A.; Hoyerová, K.; Křeček, P.; Bielach, A.; Petrášek, J.; Zhang, J.; Gaykova, V.; Stierhof, Y.-D.; et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 2009, 459, 1136–1140. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.L.; Fekete, M.L.; Klinkenberg, P.M.; Hampton, M.; Bauer, B.; Malecha, M.; Lindgren, K.; Maki, J.A.; Perera, M.A.D.N.; Nikolau, B.J.; et al. PIN6 is required for nectary auxin response and short stamen development. Plant J. 2013, 74, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Sawchuk, M.G.; Edgar, A.; Scarpella, E. Patterning of leaf vein networks by convergent auxin transport pathways. PLoS Genet. 2013, 9, e1003294. [Google Scholar] [CrossRef] [PubMed]
- Dal Bosco, C.; Dovzhenko, A.; Liu, X.; Woerner, N.; Rensch, T.; Eismann, M.; Eimer, S.; Hegermann, J.; Paponov, I.A.; Ruperti, B.; et al. The endoplasmic reticulum localized PIN8 is a pollen-specific auxin carrier involved in intracellular auxin homeostasis. Plant J. 2012, 71, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Wang, B.; Moreno, I.; Dupláková, N.; Simon, S.; Carraro, N.; Reemmer, J.; Pěnčík, A.; Chen, X.; Tejos, R.; et al. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat. Commun. 2012, 3, 941. [Google Scholar] [CrossRef] [PubMed]
- Wabnik, K.; Robert, H.S.; Smith, R.S.; Friml, J. Modeling framework for the establishment of the apical-basal embryonic axis in plants. Curr. Biol. 2013, 23, 2513–2518. [Google Scholar] [CrossRef] [PubMed]
- Dubrovsky, J.G.; Sauer, M.; Napsucialy-Mendivil, S.; Ivanchenko, M.G.; Friml, J.; Shishkova, S.; Celenza, J.; Benkova, E. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc. Natl. Acad. Sci. USA 2008, 105, 8790–8794. [Google Scholar] [CrossRef] [PubMed]
- Hay, A. ASYMMETRIC LEAVES1 and auxin activities converge to repress BREVIPEDICELLUS expression and promote leaf development in Arabidopsis. Development 2006, 133, 3955–3961. [Google Scholar] [CrossRef] [PubMed]
- Barkoulas, M.; Hay, A.; Kougioumoutzi, E.; Tsiantis, M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat. Genet. 2008, 40, 1136–1141. [Google Scholar] [CrossRef] [PubMed]
- Pernisova, M.; Klima, P.; Horak, J.; Valkova, M.; Malbeck, J.; Soucek, P.; Reichman, P.; Hoyerova, K.; Dubova, J.; Friml, J.; et al. Cytokinins modulate auxin-induced organogenesis in plants via regulation of the auxin efflux. Proc. Natl. Acad. Sci. USA 2009, 106, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Hejátko, J.; Blilou, I.; Brewer, P.B.; Friml, J.; Scheres, B.; Benková, E. In situ hybridization technique for mRNA detection in whole mount Arabidopsis samples. Nat. Protoc. 2006, 1, 1939–1946. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Paciorek, T.; Benková, E.; Friml, J. Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat. Protoc. 2006, 1, 98–103. [Google Scholar] [CrossRef] [PubMed]
- Gälweiler, L.; Guan, C.; Müller, A.; Wisman, E.; Mendgen, K.; Yephremov, A.; Palme, K. Regulation of Polar Auxin Transport by AtPIN1 in Arabidopsis Vascular Tissue. Science 1998, 282, 2226–2230. [Google Scholar] [CrossRef] [PubMed]
- Müller, A.; Guan, C.; Gälweiler, L.; Tänzler, P.; Huijser, P.; Marchant, A.; Parry, G.; Bennett, M.; Wisman, E.; Palme, K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998, 17, 6903–6911. [Google Scholar] [CrossRef] [PubMed]
- Friml, J.; Wiśniewska, J.; Benková, E.; Mendgen, K.; Palme, K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature 2002, 415, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-Regulated Auxin Transport by NRT1.1 Defines a Mechanism for Nutrient Sensing in Plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Ohmiya, A.; Hayashi, T. Immuno-gold localization of IAA in leaf cells of Prunus persica at different stages of development. Physiol. Plant. 1992, 85, 439–445. [Google Scholar] [CrossRef]
- Thomas, C.; Bronner, R.; Molinier, J.; Prinsen, E.; Van Onckelen, H.; Hahne, G. Immuno-cytochemical localization of indole-3-acetic acid during induction of somatic embryogenesis in cultured sunflower embryos. Planta 2002, 215, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Ren, Y.; Deng, Y.; Zhao, J. Auxin polar transport is essential for the development of zygote and embryo in Nicotiana tabacum L. and correlated with ABP1 and PM H+-ATPase activities. J. Exp. Bot. 2010, 61, 1853–1867. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Toyooka, K.; Sato, M.; Matsumoto, S.; Lucas, M.M.; Strnad, M.; Baluška, F.; Koshiba, T. Immunohistochemical observation of indole-3-acetic acid at the IAA synthetic maize coleoptile tips. Plant Signal. Behav. 2011, 6, 2013–2022. [Google Scholar] [CrossRef] [PubMed]
- Dong, N.; Wang, Q.; Zhang, J.; Pei, D. Immunohistochemical localization of indole-3-acetic acid during induction of adventitious root formation from cotyledon explants of walnut. J. Am. Soc. Hortic. Sci. 2011, 136, 315–319. [Google Scholar]
- Petersson, S.V.; Johansson, A.I.; Kowalczyk, M.; Makoveychuk, A.; Wang, J.Y.; Moritz, T.; Grebe, M.; Benfey, P.N.; Sandberg, G.; Ljung, K. An Auxin Gradient and Maximum in the Arabidopsis Root Apex Shown by High-Resolution Cell-Specific Analysis of IAA Distribution and Synthesis. Plant Cell 2009, 21, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Wang, B.; Zhu, J. Auxin transport during root gravitropism: Transporters and techniques. Plant Biol. 2014, 16, 50–57. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Muday, G.K. Measurement of auxin transport in Arabidopsis thaliana. Nat. Protoc. 2009, 4, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, J.; Laňková, M.; Zažímalová, E. Determination of Auxin Transport Parameters on the Cellular Level. Methods Mol. Biol. 2014, 1056, 241–253. [Google Scholar] [CrossRef] [PubMed]
- Pernet, J.J.; Pilet, P.E. Indoleacetic acid movement in the root cap. Planta 1976, 128, 183–184. [Google Scholar] [CrossRef] [PubMed]
- Davies, P.J.; Mitchell, E.K. Transport of indoleacetic acid in intact roots of Phaseolus coccineus. Planta 1972, 105, 139–154. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, E.K.; Davies, P.J. Evidence for Three Different Systems of Movement of Indoleacetic Acid in Intact Roots of Phaseolus coccineus. Physiol. Plant. 1975, 33, 290–294. [Google Scholar] [CrossRef]
- Nagao, M.; Ohwaki, Y. Auxin transport in the elongation zone of Vicia roots. Bot. Mag. Tokyo 1968, 81, 44–45. [Google Scholar] [CrossRef]
- Tsurumi, S.; Ohwaki, Y. Transport of 14C-labeled indoleacetic acid in Vicia root segments. Plant Cell Physiol. 1978, 19, 1195–1206. [Google Scholar]
- McCready, C.C. The polarity of auxin movement in segments excised from Phaseolus vulgaris L. In Biochemistry and Physiology of Plant Growth Substances; Wightman, F., Setterfield, G., Eds.; Runge Press: Ottawa, ON, Canada, 1968; pp. 1005–1023. [Google Scholar]
- Smith, C.W.; Jacobs, W.P. The movement of I4C-IAA in the hypocotyl of Phaseolus vulgaris. Am. J. Bot. 1969, 56, 492–497. [Google Scholar] [CrossRef]
- Goldsmith, M.H.M. The Polar Transport of Auxin. Annu. Rev. Plant Physiol. 1977, 28, 439–478. [Google Scholar] [CrossRef]
- Boot, K.J.M.; Hille, S.C.; Libbenga, K.R.; Peletier, L.A.; Van Spronsen, P.C.; Van Duijn, B.; Offringa, R. Modelling the dynamics of polar auxin transport in inflorescence stems of Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Ohwaki, Y.; Tsurumi, S. Auxin, transport and growth in intact roots of Vicia faba. Plant Cell Physiol. 1976, 17, 1329–1342. [Google Scholar] [CrossRef]
- Okada, K.; Ueda, J.; Komaki, M.K.; Bell, C.J.; Shimura, Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell 1991, 3, 677–684. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.M.; Brady, S.R.; Reed, R.C.; Ante, S.J.; Muday, G.K. Basipetal Auxin Transport Is Required for Gravitropism in Roots of Arabidopsis. Plant Physiol. 2000, 122, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Rashotte, A.M.; Poupart, J.; Waddell, C.S.; Muday, G.K. Transport of the two natural auxins, indole-3-butyric acid and indole-3-acetic acid, in Arabidopsis. Plant Physiol. 2003, 133, 761–772. [Google Scholar] [CrossRef] [PubMed]
- Muday, G.K.; Brady, S.R.; Argueso, C.; Deruère, J.; Kieber, J.J.; DeLong, A. RCN1-regulated phosphatase activity and EIN2 modulate hypocotyl gravitropism by a mechanism that does not require ethylene signaling. Plant Physiol. 2006, 141, 1617–1629. [Google Scholar] [CrossRef] [PubMed]
- Marchant, A.; Kargul, J.; May, S.T.; Muller, P.; Delbarre, A.; Perrot-Rechenmann, C.; Bennett, M.J. AUX1 regulates root gravitropism in Arabidopsis by facilitating auxin uptake within root apical tissues. EMBO J. 1999, 18, 2066–2073. [Google Scholar] [CrossRef] [PubMed]
- Casimiro, I.; Marchant, A.; Bhalerao, R.P.; Beeckman, T.; Dhooge, S.; Swarup, R.; Graham, N.; Inzé, D.; Sandberg, G.; Casero, P.J.; et al. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 2001, 13, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; Peer, W.A.; Taiz, L. Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 2000, 211, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Noh, B.; Bandyopadhyay, A.; Peer, W.A.; Spalding, E.P.; Murphy, A.S. Enhanced gravi- and phototropism in plant mdr mutants mislocalizing the auxin efflux protein PIN1. Nature 2003, 423, 999–1002. [Google Scholar] [CrossRef] [PubMed]
- Geisler, M.; Murphy, A.S. The ABC of auxin transport: The role of p-glycoproteins in plant development. FEBS Lett. 2006, 580, 1094–1102. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Miller, N.D.; Splitt, B.L.; Wu, G.; Spalding, E.P. Separating the Roles of Acropetal and Basipetal Auxin Transport on Gravitropism with Mutations in Two Arabidopsis Multidrug Resistance-Like ABC Transporter Genes. Plant Cell 2007, 19, 1838–1850. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Hilson, P.; Sedbrook, J.; Rosen, E.; Caspar, T.; Masson, P.H. The Arabidopsis thaliana AGRAVITROPIC 1 gene encodes a component of the polar-auxin-transport efflux carrier. Plant Biol. 1998, 95, 15112–15117. [Google Scholar] [CrossRef]
- Delbarre, A.; Muller, P.; Imhoff, V.; Guern, J. Comparison of mechanisms controlling uptake and accumulation of 2,4-dichlorophenoxy acetic acid, naphthalene-1-acetic acid, and indole-3-acetic acid in suspension-cultured tobacco cells. Planta 1996, 198, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Seifertová, D.; Skůpa, P.; Rychtář, J.; Laňková, M.; Pařezová, M.; Dobrev, P.I.; Hoyerová, K.; Petrášek, J.; Zažímalová, E. Characterization of transmembrane auxin transport in Arabidopsis suspension-cultured cells. J. Plant Physiol. 2014, 171, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Kubeš, M.; Baster, P.; Robert, S.; Dobrev, P.I.; Friml, J.; Petrášek, J.; Zažímalová, E. Defining the selectivity of processes along the auxin response chain: A study using auxin analogues. New Phytol. 2013, 200, 1034–1048. [Google Scholar] [CrossRef] [PubMed]
- Hošek, P.; Kubeš, M.; Laňková, M.; Dobrev, P.I.; Klíma, P.; Kohoutová, M.; Petrášek, J.; Hoyerová, K.; Jiřina, M.; Zažímalová, E. Auxin transport at cellular level: New insights supported by mathematical modelling. J. Exp. Bot. 2012, 63, 3815–3827. [Google Scholar] [CrossRef] [PubMed]
- Paciorek, T.; Zažímalová, E.; Ruthardt, N.; Petrášek, J.; Stierhof, Y.D.; Kleine-Vehn, J.; Morris, D.A.; Emans, N.; Jürgens, G.; Geldner, N.; et al. Auxin inhibits endocytosis and promotes its own efflux from cells. Nature 2005, 435, 1251–1256. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, J.; Elčkner, M.; Morris, D.; Zažímalová, E. Auxin efflux carrier activity and auxin accumulation regulate cell division and polarity in tobacco cells. Planta 2002, 216, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Carrier, D.J.; Abu Bakar, N.T.; Lawler, K.; Dorrian, J.M.; Haider, A.; Bennett, M.J.; Kerr, I.D. Heterologous expression of a membrane-spanning auxin importer: Implications for functional analyses of auxin transporters. Int. J. Plant Genom. 2009, 2009, 848145. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Murphy, A.S. Functional expression and characterization of Arabidopsis ABCB, AUX 1 and PIN auxin transporters in Schizosaccharomyces pombe. Plant J. 2009, 59, 179–191. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, R.; Bailly, A.; Blakeslee, J.J.; Oehring, S.C.; Vincenzetti, V.; Lee, O.R.; Paponov, I.; Palme, K.; Mancuso, S.; Murphy, A.S.; et al. Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J. Biol. Chem. 2006, 281, 30603–30612. [Google Scholar] [CrossRef] [PubMed]
- Multani, D.S.; Briggs, S.P.; Chamberlin, M.A.; Blakeslee, J.J.; Murphy, A.S.; Johal, G.S. Loss of an MDR Transporter in Compact Stalks of Maize br2 and Sorghum dw3 Mutants. Science 2003, 302, 81–84. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Hammes, U.Z.; Taylor, C.G.; Schachtman, D.P.; Nielsen, E. High-Affinity Auxin Transport by the AUX1 Influx Carrier Protein. Curr. Biol. 2006, 16, 1123–1127. [Google Scholar] [CrossRef] [PubMed]
- Blakeslee, J.J.; Murphy, A.S. Microscopic and Biochemical Visualization of Auxins in Plant Tissues. Methods Mol. Biol. 2016, 1398, 37–53. [Google Scholar] [CrossRef] [PubMed]
- Sauer, M.; Robert, S.; Kleine-Vehn, J. Auxin: Simply complicated. J. Exp. Bot. 2013, 64, 2565–2577. [Google Scholar] [CrossRef] [PubMed]
- Rigal, A.; Ma, Q.; Robert, S. Unraveling plant hormone signaling through the use of small molecules. Front. Plant Sci. 2014, 5, 373. [Google Scholar] [CrossRef] [PubMed]
- Malachowska-Ugarte, M.; Sperduto, C.; Ermolovich, Y.V.; Sauchuk, A.L.; Jurášek, M.; Litvinovskaya, R.P.; Straltsova, D.; Smolich, I.; Zhabinskii, V.N.; Drašar, P.; et al. Brassinosteroid-BODIPY conjugates: Design, synthesis, and properties. Steroids 2015, 102, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Prandi, C.; Ghigo, G.; Occhiato, E.G.; Scarpi, D.; Begliomini, S.; Lace, B.; Alberto, G.; Artuso, E.; Blangetti, M. Tailoring fluorescent strigolactones for in vivo investigations: A computational and experimental study. Org. Biomol. Chem. 2014, 12, 2960–2968. [Google Scholar] [CrossRef] [PubMed]
- Enciso, A.E.; Garzoni, M.; Pavan, G.M.; Simanek, E.E. Influence of linker groups on the solubility of triazine dendrimers. New J. Chem. 2015, 39, 1247–1252. [Google Scholar] [CrossRef]
- Bieleszová, K.; Pařízková, B.; Kubeš, M.; Husičková, A.; Kubala, M.; Sedlářová, M.; Doležal, K.; Strnad, M.; Novák, O.; Žukauskaite, A. New fluorescently labeled auxins exhibit promising anti-auxin activity. N. Biotechnol. 2017. Under review. [Google Scholar]
- Lace, B.; Prandi, C. Shaping Small Bioactive Molecules to Untangle Their Biological Function: A Focus on Fluorescent Plant Hormones. Mol. Plant 2016, 9, 1099–1118. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhu, J.J. Quantum dots for fluorescent biosensing and bio-imaging applications. Analyst 2013, 138, 2506–2515. [Google Scholar] [CrossRef] [PubMed]
- Wegner, K.D.; Hildebrandt, N. Quantum dots: Bright and versatile in vitro and in vivo fluorescence imaging biosensors. Chem. Soc. Rev. 2015, 44, 4792–4834. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Wei, J.; Liao, Q.; Yu, Y.; Lin, B. In situ fluorescence labelling of jasmonic acid binding sites in plant tissues with cadmium-free quantum dots. IET Nanobiotechnol. 2015, 9, 35–42. [Google Scholar] [CrossRef]
- Lavis, L.D.; Thomas, J.; Rutkoski, A.; Ronald, T. Raines Tuning the pKa of Fluorescein to Optimize Binding Assays. Anal. Chem. 2007, 79, 6775–6782. [Google Scholar] [CrossRef] [PubMed]
- Panchuk-Voloshina, N.; Haugland, R.P.; Bishop-Stewart, J.; Bhalgat, M.K.; Millard, P.J.; Mao, F.; Leung, W.-Y.; Haugland, R.P. Alexa Dyes, a Series of New Fluorescent Dyes that Yield Exceptionally Bright, Photostable Conjugates. J. Histochem. Cytochem. 1999, 47, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Lavis, L.D.; Raines, R.T. Bright Ideas for Chemical Biology. ACS Chem. Biol. 2008, 3, 142–155. [Google Scholar] [CrossRef] [PubMed]
- Beija, M.; Afonso, C.A.M.; Martinho, J.M.G. Synthesis and applications of Rhodamine derivatives as fluorescent probes. Chem. Soc. Rev. 2009, 38, 2410–2433. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, K.-I.; Nakamura, S.; Fukunaga, S.; Nishimura, T.; Jenness, M.K.; Murphy, A.S.; Motose, H.; Nozaki, H.; Furutani, M.; Aoyama, T. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc. Natl. Acad. Sci. USA 2014, 111, 11557–11562. [Google Scholar] [CrossRef] [PubMed]
- Muir, R.M.; Fujita, T.; Hansch, C. Structure-activity relationship in the auxin activity of mono-substituted phenylacetic acids. Plant Physiol. 1967, 42, 1519–1526. [Google Scholar] [CrossRef] [PubMed]
- Kaethner, T.M. Conformational change theory for auxin structure-activity relationships. Nature 1977, 267, 19–23. [Google Scholar] [CrossRef]
- Calderon-Villalobos, L.I.; Tan, X.; Zheng, N.; Estelle, M. Auxin Perception-Structural Insights. Cold Spring Harb. Perspect. Biol. 2010, 2, a005546. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, K. Auxin herbicides: Current status of mechanism and mode of action. Pest Manag. Sci. 2010, 66, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Muscolo, A.; Sidari, M.; Francioso, O.; Tugnoli, V.; Nardi, S. The auxin-like activity of humic substances is related to membrane interactions in carrot cell cultures. J. Chem. Ecol. 2007, 33, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Sokołowska, K.; Kizińska, J.; Szewczuk, Z.; Banasiak, A. Auxin conjugated to fluorescent dyes—A tool for the analysis of auxin transport pathways. Plant Biol. 2014, 16, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Tsuda, E.; Yang, H.; Nishimura, T.; Uehara, Y.; Sakai, T.; Furutani, M.; Koshiba, T.; Hirose, M.; Nozaki, H.; Murphy, A.S.; et al. Alkoxy-auxins are selective inhibitors of auxin transport mediated by PIN, ABCB, and AUX1 transporters. J. Biol. Chem. 2011, 286, 2354–2364. [Google Scholar] [CrossRef] [PubMed]
- Mancuso, S.; Marras, A.M.; Magnus, V.; Baluška, F. Noninvasive and continuous recordings of auxin fluxes in intact root apex with a carbon nanotube-modified and self-referencing microelectrode. Anal. Biochem. 2005, 341, 344–351. [Google Scholar] [CrossRef] [PubMed]
- McLamore, E.S.; Diggs, A.; Calvo Marzal, P.; Shi, J.; Blakeslee, J.J.; Peer, W.A.; Murphy, A.S.; Porterfield, D.M. Non-invasive quantification of endogenous root auxin transport using an integrated flux microsensor technique. Plant J. 2010, 63, 1004–1016. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, H.; Dhonukshe, P.; Brewer, P.B.; Friml, J. Spatiotemporal asymmetric auxin distribution: A means to coordinate plant development. Cell. Mol. Life Sci. 2006, 63, 2738–2754. [Google Scholar] [CrossRef] [PubMed]
- Grahm, L. Measurements of Geoelectric and Anxin-Induced Potentials in Coleoptiles with a Refined Vibrating Electrode Technique. Physiol. Plant. 1964, 17, 231–261. [Google Scholar] [CrossRef]
- Johnsson, A. Photoinduced Lateral Potentials in Zea mays. Physiol. Plant. 1965, 18, 574–576. [Google Scholar] [CrossRef]
- Bailly, A.; Sovero, V.; Vincenzetti, V.; Santelia, D.; Bartnik, D.; Koenig, B.W.; Mancuso, S.; Martinoia, E.; Geisler, M. Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J. Biol. Chem. 2008, 283, 21817–21826. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-Y.; Henrichs, S.; Bailly, A.; Vincenzetti, V.; Sovero, V.; Mancuso, S.; Pollmann, S.; Kim, D.; Geisler, M.; Nam, H.-G. Identification of an ABCB/P-glycoprotein-specific inhibitor of auxin transport by chemical genomics. J. Biol. Chem. 2010, 285, 23309–23317. [Google Scholar] [CrossRef] [PubMed]
- Santelia, D.; Vincenzetti, V.; Azzarello, E.; Bovet, L.; Fukao, Y.; Düchtig, P.; Mancuso, S.; Martinoia, E.; Geisler, M. MDR-like ABC transporter AtPGP4 is involved in auxin-mediated lateral root and root hair development. FEBS Lett. 2005, 579, 5399–5406. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Bailly, A.; Zwiewka, M.; Henrichs, S.; Azzarello, E.; Mancuso, S.; Maeshima, M.; Friml, J.; Schulz, A.; Geisler, M. Arabidopsis TWISTED DWARF1 functionally interacts with auxin exporter ABCB1 on the root plasma membrane. Plant Cell 2013, 25, 202–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlicht, M.; Strnad, M.; Scanlon, M.J.; Mancuso, S.; Hochholdinger, F.; Palme, K.; Volkmann, D.; Menzel, D.; Baluska, F. Auxin immunolocalization implicates vesicular neurotransmitter-like mode of polar auxin transport in root apices. Plant Signal. Behav. 2006, 1, 122–133. [Google Scholar] [CrossRef] [PubMed]
- Knöller, A.S.; Blakeslee, J.J.; Richards, E.L.; Peer, W.A.; Murphy, A.S. Brachytic2/ZmABCB1 functions in IAA export from intercalary meristems. J. Exp. Bot. 2010, 61, 3689–3696. [Google Scholar] [CrossRef] [PubMed]
- Sadanandom, A.; Napier, R.M. Biosensors in plants. Curr. Opin. Plant Biol. 2010, 13, 736–743. [Google Scholar] [CrossRef] [PubMed]
- Novák, O.; Pěnčík, A.; Ljung, K. Identification and Profiling of Auxin and Auxin Metabolites. In Auxin and Its Role in Plant Development, 1st ed.; Zažímalová, E., Petrášek, J., Benková, E., Eds.; Springer: London, UK, 2014; Volume 33, pp. 39–60. ISBN 978-3-7091-1526-8. [Google Scholar]
- Porfírio, S.; Gomes da Silva, M.D.R.; Peixe, A.; Cabrita, M.J.; Azadi, P. Current analytical methods for plant auxin quantification—A review. Anal. Chim. Acta 2016, 902, 8–21. [Google Scholar] [CrossRef] [PubMed]
- Novák, O.; Napier, R.; Ljung, K. Zooming In on Plant Hormone Analysis: Tissue- and Cell-Specific Approaches. Annu. Rev. Plant Biol. 2017, 68, 323–348. [Google Scholar] [CrossRef] [PubMed]
- Tarkowská, D.; Novák, O.; Floková, K.; Tarkowski, P.; Turečková, V.; Grúz, J.; Rolčík, J.; Strnad, M. Quo vadis plant hormone analysis? Planta 2014, 240, 55–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Hegeman, A.D.; Gardner, G.; Cohen, J.D. Protocol: High-throughput and quantitative assays of auxin and auxin precursors from minute tissue samples. Plant Methods 2012, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Novák, O.; Hényková, E.; Sairanen, I.; Kowalczyk, M.; Pospíšil, T.; Ljung, K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012, 72, 523–536. [Google Scholar] [CrossRef] [PubMed]
- Uggla, C.; Moritz, T.; Sandberg, G.; Sundberg, B. Auxin as a positional signal in pattern formation in plants. Proc. Natl. Acad. Sci. USA 1996, 93, 9282–9286. [Google Scholar] [CrossRef] [PubMed]
- Immanen, J.; Nieminen, K.; Smolander, O.P.; Kojima, M.; Alonso Serra, J.; Koskinen, P.; Zhang, J.; Elo, A.; Mähönen, A.P.; Street, N.; et al. Cytokinin and Auxin Display Distinct but Interconnected Distribution and Signaling Profiles to Stimulate Cambial Activity. Curr. Biol. 2016, 26, 1990–1997. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wang, R.-S.; Zhu, M.; Jeon, B.W.; Albert, R.; Chen, S.; Assmann, S.M. Abscisic Acid-Responsive Guard Cell Metabolomes of Arabidopsis Wild-Type and gpa1 G-Protein Mutants. Plant Cell 2013, 25, 4789–4811. [Google Scholar] [CrossRef] [PubMed]
- Bargmann, B.O.R.; Vanneste, S.; Krouk, G.; Nawy, T.; Efroni, I.; Shani, E.; Choe, G.; Friml, J.; Bergmann, D.C.; Estelle, M.; et al. A map of cell type-specific auxin responses. Mol. Syst. Biol. 2014, 9, 688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rosquete, R.M.; Barbez, E.; Kleine-Vehn, J. Cellular auxin homeostasis: Gatekeeping is housekeeping. Mol. Plant 2012, 5, 772–786. [Google Scholar] [CrossRef] [PubMed]
- Uslu, V.V.; Grossmann, G. The biosensor toolbox for plant developmental biology. Curr. Opin. Plant Biol. 2016, 29, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Z.; Wang, M.; Meng, X.; Yin, H. Electrochemical immunoassay platform for high sensitivity detection of indole-3-acetic acid. Electrochim. Acta 2013, 96, 66–73. [Google Scholar] [CrossRef]
- Yin, H.; Xu, Z.; Zhou, Y.; Wang, M.; Ai, S. An ultrasensitive electrochemical immunosensor platform with double signal amplification for indole-3-acetic acid determinations in plant seeds. Analyst 2013, 138, 1851–1857. [Google Scholar] [CrossRef] [PubMed]
- Sun, B.; Chen, L.; Xu, Y.; Liu, M.; Yin, H.; Ai, S. Ultrasensitive photoelectrochemical immunoassay of indole-3-acetic acid based on the MPA modified CdS/RGO nanocomposites decorated ITO electrode. Biosens. Bioelectron. 2014, 51, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wu, Z.-Y.; Xiao, L.-T.; Zeng, G.-M.; Huang, G.-H.; Shen, G.-L.; Yu, R.-Q. A novel piezoelectric biosensor for the detection of phytohormone beta-indole acetic acid. Anal. Sci. 2002, 18, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Zhou, H.; Chen, C.; Li, Z.; Zhou, J. On-Line Monitoring 1H-Indole-3-Acetic Acid in Plant Tissues Using Molecular Imprinting Monolayer Techniques on a Surface Plasmon Resonance Sensor. Anal. Lett. 2011, 44, 2911–2921. [Google Scholar] [CrossRef]
- Li, J.; Yin, W.; Tan, Y.; Pan, H. A sensitive electrochemical molecularly imprinted sensor based on catalytic amplification by silver nanoparticles for 3-indoleacetic acid determination. Sens. Actuators B Chem. 2014, 197, 109–115. [Google Scholar] [CrossRef]
- Hernández, P.; Galán, F.; Nieto, O.; Hernández, L. Direct determination of indole-3-acetic acid in plant tissues by electroanalytical techniques using a carbon paste modified with OV-17 electrode. Electroanalysis 1994, 6, 577–583. [Google Scholar] [CrossRef]
- Wu, K.; Sun, Y.; Hu, S. Development of an amperometric indole-3-acetic acid sensor based on carbon nanotubes film coated glassy carbon electrode. Sens. Actuators B Chem. 2003, 96, 658–662. [Google Scholar] [CrossRef]
- Sandberg, G. Presence of indole-3-acetic acid in chloroplasts of Nicotiana tabacum and Pinus sylvestris. Planta 1990, 180, 562–568. [Google Scholar] [CrossRef] [PubMed]
- Polanská, L.; Vičánková, A.; Nováková, M.; Malbeck, J.; Dobrev, P.I.; Brzobohatý, B.; Vaňková, R.; Macháčková, I. Altered cytokinin metabolism affects cytokinin, auxin, and abscisic acid contents in leaves and chloroplasts, and chloroplast ultrastructure in transgenic tobacco. J. Exp. Bot. 2007, 58, 637–649. [Google Scholar] [CrossRef] [PubMed]
- Wells, D.M.; Laplaze, L.; Bennett, M.J.; Vernoux, T. Biosensors for phytohormone quantification: Challenges, solutions, and opportunities. Trends Plant Sci. 2013, 18, 244–249. [Google Scholar] [CrossRef] [PubMed]
- Okumoto, S.; Jones, A.; Frommer, W.B. Quantitative Imaging with Fluorescent Biosensors. Annu. Rev. Plant Biol. 2012, 63, 663–706. [Google Scholar] [CrossRef] [PubMed]
- Reddy, G.V.; Gordon, S.P.; Meyerowitz, E.M. Unravelling developmental dynamics: Transient intervention and live imaging in plants. Nat. Rev. Mol. Cell Biol. 2007, 8, 491–501. [Google Scholar] [CrossRef] [PubMed]
- Kirchhelle, C.; Moore, I. A Simple Chamber for Long-term Confocal Imaging of Root and Hypocotyl Development. J. Vis. Exp. 2017, 123, 55331. [Google Scholar] [CrossRef] [PubMed]
- Von Wangenheim, D.; Hauschild, R.; Fendrych, M.; Barone, V.; Friml, J. Live Tracking of Moving Samples in Confocal Microscopy for Vertically Grown Plant Roots. eLife 2017, 6, e26792. [Google Scholar] [CrossRef] [PubMed]
- Maizel, A.; von Wangenheim, D.; Federici, F.; Haseloff, J.; Stelzer, E.H.K. High-resolution live imaging of plant growth in near physiological bright conditions using light sheet fluorescence microscopy. Plant J. 2011, 68, 377–385. [Google Scholar] [CrossRef] [PubMed]
- Ovečka, M.; Vaškebová, L.; Komis, G.; Luptovčiak, I.; Smertenko, A.; Šamaj, J. Preparation of plants for developmental and cellular imaging by light-sheet microscopy. Nat. Protoc. 2015, 10, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, G.; Guo, W.-J.; Ehrhardt, D.W.; Frommer, W.B.; Sit, R.V.; Quake, S.R.; Meier, M. The RootChip: An Integrated Microfluidic Chip for Plant Science. Plant Cell 2011, 23, 4234–4240. [Google Scholar] [CrossRef] [PubMed]
- Grossmann, G.; Meier, M.; Cartwright, H.N.; Sosso, D.; Quake, S.R.; Ehrhardt, D.W.; Frommer, W.B. Time-lapse Fluorescence Imaging of Arabidopsis Root Growth with Rapid Manipulation of The Root Environment Using The RootChip. J. Vis. Exp. 2012, 65, 4290. [Google Scholar] [CrossRef] [PubMed]
- Evans, C.L.; Xie, X.S. Coherent anti-Stokes Raman scattering microscopy: Chemical imaging for biology and medicine. Annu. Rev. Anal. Chem. 2008, 1, 883–909. [Google Scholar] [CrossRef] [PubMed]
- Freudiger, C.W.; Min, W.; Saar, B.G.; Lu, S.; Holtom, G.R.; Tsai, J.C.; Kang, J.X.; Xie, X.S. Label-Free Biomedical Imaging with High Sensitivity by Stimulated Raman Scattering Microscopy. Science 2013, 322, 1857–1861. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pařízková, B.; Pernisová, M.; Novák, O. What Has Been Seen Cannot Be Unseen—Detecting Auxin In Vivo. Int. J. Mol. Sci. 2017, 18, 2736. https://doi.org/10.3390/ijms18122736
Pařízková B, Pernisová M, Novák O. What Has Been Seen Cannot Be Unseen—Detecting Auxin In Vivo. International Journal of Molecular Sciences. 2017; 18(12):2736. https://doi.org/10.3390/ijms18122736
Chicago/Turabian StylePařízková, Barbora, Markéta Pernisová, and Ondřej Novák. 2017. "What Has Been Seen Cannot Be Unseen—Detecting Auxin In Vivo" International Journal of Molecular Sciences 18, no. 12: 2736. https://doi.org/10.3390/ijms18122736
APA StylePařízková, B., Pernisová, M., & Novák, O. (2017). What Has Been Seen Cannot Be Unseen—Detecting Auxin In Vivo. International Journal of Molecular Sciences, 18(12), 2736. https://doi.org/10.3390/ijms18122736