Evaluation of the Expression of Amine Oxidase Proteins in Breast Cancer
Abstract
:1. Introduction
2. Results
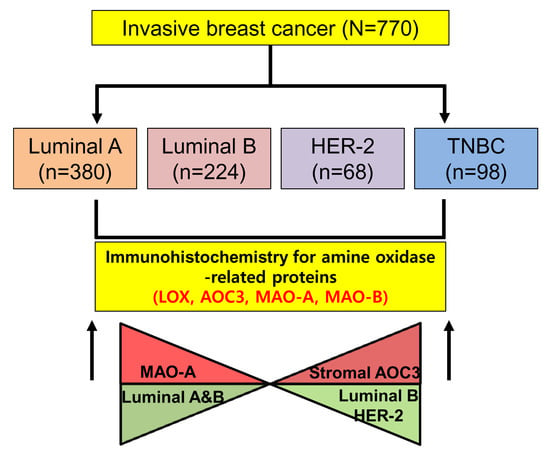
2.1. Patient Characteristics
2.2. Differential Expression of Amine Oxidase in Different Tumor Subtypes
2.3. Correlation of the Expression of Amine Oxidase Proteins in Breast Cancer
2.4. Correlation between the Expression of Amine Oxidase and Clinicopathological Characteristics
2.5. Functional Analysis Using STRING Database
2.6. Effect of the Expression of Amine Oxidase on Survival
3. Discussion
4. Materials and Methods
4.1. Patient Selection and Histological Evaluation
4.2. Tissue Microarray
4.3. Immunohistochemistry
4.4. Interpretation of Immunohistochemical Staining
4.5. Tumor Phenotype Classification
4.6. Functional Analysis Using STRING Database
4.7. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mondovi, B.; Finazzi Agro, A. Structure and function of amine oxidase. Adv. Exp. Med. Biol. 1982, 148, 141–153. [Google Scholar] [PubMed]
- Kumar, V.; Dooley, D.M.; Freeman, H.C.; Guss, J.M.; Harvey, I.; McGuirl, M.A.; Wilce, M.C.; Zubak, V.M. Crystal structure of a eukaryotic (pea seedling) copper-containing amine oxidase at 2.2 A resolution. Structure 1996, 4, 943–955. [Google Scholar] [CrossRef]
- Reynaud, C.; Ferreras, L.; Di Mauro, P.; Kan, C.; Croset, M.; Bonnelye, E.; Pez, F.; Thomas, C.; Aimond, G.; Karnoub, A.E.; et al. Lysyl oxidase is a strong determinant of tumor cell colonization in bone. Cancer Res. 2017, 77, 268–278. [Google Scholar] [CrossRef] [PubMed]
- Kostoro, J.; Chang, S.J.; Clark Lai, Y.C.; Wu, C.C.; Chai, C.Y.; Kwan, A.L. Overexpression of vascular adhesion protein-1 is associated with poor prognosis of astrocytomas. APMIS 2016, 124, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Toiyama, Y.; Miki, C.; Inoue, Y.; Kawamoto, A.; Kusunoki, M. Circulating form of human vascular adhesion protein-1 (VAP-1): Decreased serum levels in progression of colorectal cancer and predictive marker of lymphatic and hepatic metastasis. J. Surg. Oncol. 2009, 99, 368–372. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, H.; Toiyama, Y.; Ohi, M.; Mohri, Y.; Miki, C.; Kusunoki, M. Serum soluble vascular adhesion protein-1 is a valuable prognostic marker in gastric cancer. J. Surg. Oncol. 2011, 103, 695–699. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.B.; Shao, C.; Li, X.; Li, Q.; Hu, P.; Shi, C.; Li, Y.; Chen, Y.T.; Yin, F.; Liao, C.P.; et al. Monoamine oxidase a mediates prostate tumorigenesis and cancer metastasis. J. Clin. Investig. 2014, 124, 2891–2908. [Google Scholar] [CrossRef] [PubMed]
- Sharpe, M.A.; Baskin, D.S. Monoamine oxidase b levels are highly expressed in human gliomas and are correlated with the expression of hif-1α and with transcription factors Sp1 and Sp3. Oncotarget 2016, 7, 3379–3393. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.E.; Jung, W.H.; Koo, J.S. Molecules involved in epithelial-mesenchymal transition and epithelial-stromal interaction in phyllodes tumors: Implications for histologic grade and prognosis. Tumour Biol. 2012, 33, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Reis-Filho, J.S.; Tutt, A.N. Triple negative tumours: A critical review. Histopathology 2008, 52, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, C.; Sousa, B.; Albergaria, A.; Paredes, J.; Dufloth, R.; Vieira, D.; Schmitt, F.; Baltazar, F. GLUT1 and CAIX expression profiles in breast cancer correlate with adverse prognostic factors and MCT1 overexpression. Histol. Histopathol. 2011, 26, 1279–1286. [Google Scholar] [PubMed]
- Choi, J.; Jung, W.H.; Koo, J.S. Metabolism-related proteins are differentially expressed according to the molecular subtype of invasive breast cancer defined by surrogate immunohistochemistry. Pathobiology 2013, 80, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kim do, H.; Jung, W.H.; Koo, J.S. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr. Relat. Cancer 2013, 20, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Hsia, L.T.; Ashley, N.; Ouaret, D.; Wang, L.M.; Wilding, J.; Bodmer, W.F. Myofibroblasts are distinguished from activated skin fibroblasts by the expression of AOC3 and other associated markers. Proc. Natl. Acad. Sci. USA 2016, 113, E2162–E2171. [Google Scholar] [CrossRef] [PubMed]
- Mellone, M.; Hanley, C.J.; Thirdborough, S.; Mellows, T.; Garcia, E.; Woo, J.; Tod, J.; Frampton, S.; Jenei, V.; Moutasim, K.A.; et al. Induction of fibroblast senescence generates a non-fibrogenic myofibroblast phenotype that differentially impacts on cancer prognosis. Aging 2016, 9, 114–132. [Google Scholar] [CrossRef] [PubMed]
- Hanley, C.J.; Noble, F.; Ward, M.; Bullock, M.; Drifka, C.; Mellone, M.; Manousopoulou, A.; Johnston, H.E.; Hayden, A.; Thirdborough, S.; et al. A subset of myofibroblastic cancer-associated fibroblasts regulate collagen fiber elongation, which is prognostic in multiple cancers. Oncotarget 2016, 7, 6159–6174. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Kim, H.M.; Koo, J.S. Differential expression of cancer-associated fibroblast-related proteins according to molecular subtype and stromal histology in breast cancer. Breast Cancer Res. Treat. 2015, 149, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Tchou, J.; Kossenkov, A.V.; Chang, L.; Satija, C.; Herlyn, M.; Showe, L.C.; Pure, E. Human breast cancer associated fibroblasts exhibit subtype specific gene expression profiles. BMC Med. Genom. 2012, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Akagawa, S.; Ohuchida, K.; Torata, N.; Hattori, M.; Eguchi, D.; Fujiwara, K.; Kozono, S.; Cui, L.; Ikenaga, N.; Ohtsuka, T.; et al. Peritoneal myofibroblasts at metastatic foci promote dissemination of pancreatic cancer. Int. J. Oncol. 2014, 45, 113–120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lizcano, J.M.; Escrich, E.; Ribalta, T.; Muntane, J.; Unzeta, M. Amine oxidase activities in rat breast cancer induced experimentally with 7,12-dimethylbenz(α)anthracene. Biochem. Pharmacol. 1991, 42, 263–269. [Google Scholar] [CrossRef]
- Lizcano, J.M.; Escrich, E.; Tipton, K.F.; Unzeta, M. Amine oxidase activities in chemically-induced mammary cancer in the rat. J. Neural Transm. Suppl. 1990, 32, 323–326. [Google Scholar] [PubMed]
- Ren, Y.; Jiang, H.; Ma, D.; Nakaso, K.; Feng, J. Parkin degrades estrogen-related receptors to limit the expression of monoamine oxidases. Hum. Mol. Genet. 2011, 20, 1074–1083. [Google Scholar] [CrossRef] [PubMed]
- Rybaczyk, L.A.; Bashaw, M.J.; Pathak, D.R.; Huang, K. An indicator of cancer: Downregulation of monoamine oxidase-A in multiple organs and species. BMC Genom. 2008, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, K.; Shih, J.C.; Teng, C.T. Estrogen-related receptors-stimulated monoamine oxidase B promoter activity is down-regulated by estrogen receptors. Mol. Endocrinol. 2006, 20, 1547–1561. [Google Scholar] [CrossRef] [PubMed]
- Willy, P.J.; Murray, I.R.; Qian, J.; Busch, B.B.; Stevens, W.C., Jr.; Martin, R.; Mohan, R.; Zhou, S.; Ordentlich, P.; Wei, P.; et al. Regulation of PPARγ coactivator 1α (PGC-1α) signaling by an estrogen-related receptor α (ERRα) ligand. Proc. Natl. Acad. Sci. USA 2004, 101, 8912–8917. [Google Scholar] [CrossRef] [PubMed]
- Wuest, M.; Kuchar, M.; Sharma, S.K.; Richter, S.; Hamann, I.; Wang, M.; Vos, L.; Mackey, J.R.; Wuest, F.; Loser, R. Targeting lysyl oxidase for molecular imaging in breast cancer. Breast Cancer Res. 2015, 17, 107. [Google Scholar] [CrossRef] [PubMed]
- Kirschmann, D.A.; Seftor, E.A.; Fong, S.F.; Nieva, D.R.; Sullivan, C.M.; Edwards, E.M.; Sommer, P.; Csiszar, K.; Hendrix, M.J. A molecular role for lysyl oxidase in breast cancer invasion. Cancer Res. 2002, 62, 4478–4483. [Google Scholar] [PubMed]
- Xiao, Q.; Ge, G. Lysyl oxidase, extracellular matrix remodeling and cancer metastasis. Cancer Microenviron. 2012, 5, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Shaheed, S.U.; Rustogi, N.; Scally, A.; Wilson, J.; Thygesen, H.; Loizidou, M.A.; Hadjisavvas, A.; Hanby, A.; Speirs, V.; Loadman, P.; et al. Identification of stage-specific breast markers using quantitative proteomics. J. Proteome Res. 2013, 12, 5696–5708. [Google Scholar] [CrossRef] [PubMed]
- Marttila-Ichihara, F.; Auvinen, K.; Elima, K.; Jalkanen, S.; Salmi, M. Vascular adhesion protein-1 enhances tumor growth by supporting recruitment of Gr-1+CD11b+ myeloid cells into tumors. Cancer Res. 2009, 69, 7875–7883. [Google Scholar] [CrossRef] [PubMed]
- Reymond, N.; d’Agua, B.B.; Ridley, A.J. Crossing the endothelial barrier during metastasis. Nat. Rev. Cancer 2013, 13, 858–870. [Google Scholar] [CrossRef] [PubMed]
- Kurkijarvi, R.; Adams, D.H.; Leino, R.; Mottonen, T.; Jalkanen, S.; Salmi, M. Circulating form of human vascular adhesion protein-1 (VAP-1): Increased serum levels in inflammatory liver diseases. J. Immunol. 1998, 161, 1549–1557. [Google Scholar] [PubMed]
- Barker, H.E.; Cox, T.R.; Erler, J.T. The rationale for targeting the lox family in cancer. Nat. Rev. Cancer 2012, 12, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Tu, S.H.; Huang, C.S.; Chen, C.S.; Ho, C.T.; Lin, H.W.; Lee, C.H.; Chang, H.W.; Chang, C.H.; Wu, C.H.; et al. Human breast cancer cell metastasis is attenuated by lysyl oxidase inhibitors through down-regulation of focal adhesion kinase and the paxillin-signaling pathway. Breast Cancer Res. Treat. 2012, 134, 989–1004. [Google Scholar] [CrossRef] [PubMed]
- Kanapathipillai, M.; Mammoto, A.; Mammoto, T.; Kang, J.H.; Jiang, E.; Ghosh, K.; Korin, N.; Gibbs, A.; Mannix, R.; Ingber, D.E. Inhibition of mammary tumor growth using lysyl oxidase-targeting nanoparticles to modify extracellular matrix. Nano Lett. 2012, 12, 3213–3217. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, M.; Adamo, H.; Bergh, A.; Halin Bergstrom, S. Inhibition of lysyl oxidase and lysyl oxidase-like enzymes has tumour-promoting and tumour-suppressing roles in experimental prostate cancer. Sci. Rep. 2016, 6, 19608. [Google Scholar] [CrossRef] [PubMed]
- Kushal, S.; Wang, W.; Vaikari, V.P.; Kota, R.; Chen, K.; Yeh, T.S.; Jhaveri, N.; Groshen, S.L.; Olenyuk, B.Z.; Chen, T.C.; et al. Monoamine oxidase A (MAO A) inhibitors decrease glioma progression. Oncotarget 2016, 7, 13842–13853. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.T.; Choi, M.R.; Doh, M.S.; Jung, K.H.; Chai, Y.G. Effects of the monoamine oxidase inhibitors pargyline and tranylcypromine on cellular proliferation in human prostate cancer cells. Oncol. Rep. 2013, 30, 1587–1592. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Adisetiyo, H.; Tamura, S.; Grande, F.; Garofalo, A.; Roy-Burman, P.; Neamati, N. Dual inhibition of survivin and MAOA synergistically impairs growth of PTEN-negative prostate cancer. Br. J. Cancer 2015, 113, 242–251. [Google Scholar] [CrossRef] [PubMed]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Hammond, M.E.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American society of clinical oncology/college of american pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef] [PubMed]
- Wolff, A.C.; Hammond, M.E.; Schwartz, J.N.; Hagerty, K.L.; Allred, D.C.; Cote, R.J.; Dowsett, M.; Fitzgibbons, P.L.; Hanna, W.M.; Langer, A.; et al. American society of clinical oncology/college of american pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J. Clin. Oncol. 2007, 25, 118–145. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Jung, W.H.; Koo, J.S. Clinicopathologic features of molecular subtypes of triple negative breast cancer based on immunohistochemical markers. Histol. Histopathol. 2012, 27, 1481–1493. [Google Scholar] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thurlimann, B.; Senn, H.J. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the st. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]





| Parameter | Total (n = 770) (%) | Luminal A (n = 380) (%) | Luminal B (n = 224) (%) | HER-2 (n = 68) (%) | TNBC (n = 98) (%) | p-Value |
|---|---|---|---|---|---|---|
| Age (years) | 0.040 | |||||
| ≤50 | 463 (60.1) | 234 (61.6) | 144 (64.3) | 32 (47.1) | 53 (54.1) | |
| >50 | 307 (39.9) | 146 (38.4) | 80 (35.7) | 36 (52.9) | 45 (45.9) | |
| Histological grade | <0.001 | |||||
| I/II | 559 (72.6) | 354 (93.2) | 135 (60.3) | 37 (54.4) | 33 (33.7) | |
| III | 211 (27.4) | 26 (6.8) | 89 (39.7) | 31 (45.6) | 65 (66.3) | |
| Tumor stage | 0.158 | |||||
| T1 | 433 (56.2) | 228 (60.0) | 122 (54.5) | 35 (51.5) | 48 (49.0) | |
| T2/T3 | 337 (43.8) | 152 (40.0) | 102 (45.5) | 33 (48.5) | 50 (51.0) | |
| Nodal metastasis | 0.028 | |||||
| Absent | 460 (59.7) | 213 (56.1) | 133 (59.4) | 43 (63.2) | 71 (72.4) | |
| Present | 310 (40.3) | 167 (43.9) | 91 (40.6) | 25 (36.8) | 27 (27.6) | |
| Estrogen-receptor status | <0.001 | |||||
| Negative | 196 (25.5) | 9 (2.4) | 21 (9.4) | 68 (100.0) | 98 (100.0) | |
| Positive | 574 (74.5) | 371 (97.6) | 203 (90.6) | 0 (0.0) | 0 (0.0) | |
| Progesterone-receptor status | <0.001 | |||||
| Negative | 263 (34.2) | 48 (12.6) | 49 (21.9) | 68 (100.0) | 98 (100.0) | |
| Positive | 507 (65.8) | 332 (87.4) | 175 (78.1) | 0 (0.0) | 0 (0.0) | |
| HER-2 status | <0.001 | |||||
| Negative | 609 (79.1) | 380 (100.0) | 131 (58.5) | 0 (0.0) | 98 (100.0) | |
| Positive | 161 (20.9) | 0 (0.0) | 93 (41.5) | 68 (100.0) | 0 (0.0) | |
| Ki-67 LI (%) | <0.001 | |||||
| ≤14 | 457 (59.4) | 380 (100.0) | 49 (21.9) | 17 (25.0) | 11 (11.2) | |
| >14 | 313 (40.6) | 0 (0.0) | 175 (78.1) | 51 (75.0) | 87 (88.8) | |
| Duration of clinical follow-up (months, mean ± SD) | 71.8 ± 21.7 | 72.9 ± 20.1 | 71.6 ± 21.6 | 64.1 ± 25.3 | 73.5 ± 24.4 | 0.019 |
| Parameter | Total (n = 770) (100%) | Luminal A (n = 380) (49.4%) | Luminal B (n = 224) (29.1%) | HER-2 (n = 68) (8.8%) | TNBC (n = 98) (12.7%) | p-Value |
|---|---|---|---|---|---|---|
| LOX | 0.178 | |||||
| Negative | 413 (53.6) | 218 (57.4) | 113 (50.4) | 31 (45.6) | 51 (52.0) | |
| Positive | 357 (46.4) | 162 (42.6) | 111 (49.6) | 37 (54.4) | 47 (48.0) | |
| AOC3 | 0.199 | |||||
| Negative | 178 (23.1) | 84 (22.1) | 49 (21.9) | 14 (20.6) | 31 (31.6) | |
| Positive | 592 (76.9) | 296 (77.9) | 175 (78.1) | 54 (79.4) | 67 (68.4) | |
| AOC3 (S) | <0.001 | |||||
| Negative | 681 (88.4) | 352 (92.6) | 182 (81.3) | 58 (85.3) | 89 (90.8) | |
| Positive | 89 (11.6) | 28 (7.4) | 42 (18.8) | 10 (14.7) | 9 (9.2) | |
| MAOA | <0.001 | |||||
| Negative | 546 (70.6) | 229 (60.3) | 168 (75.0) | 59 (86.8) | 90 (91.8) | |
| Positive | 224 (29.1) | 151 (39.7) | 56 (25.0) | 9 (13.2) | 8 (8.2) | |
| MAOB | 0.020 | |||||
| Negative | 658 (85.5) | 337 (88.7) | 189 (84.4) | 57 (83.8) | 75 (76.5) | |
| Positive | 112 (14.5) | 43 (11.3) | 35 (15.6) | 11 (16.2) | 23 (23.5) | |
| MAOB (S) | 0.075 | |||||
| Negative | 648 (84.2) | 331 (87.1) | 178 (79.5) | 55 (80.9) | 84 (85.7) | |
| Positive | 122 (15.8) | 49 (12.9) | 46 (20.5) | 13 (19.1) | 14 (14.3) |
| Parameters | LOX | AOC3 | AOC3 (S) | MAOA | MAOB | MAOB (S) |
|---|---|---|---|---|---|---|
| LOX | ||||||
| Correlation coefficient | 0.238 | 0.144 | 0.001 | 0.060 | 0.160 | |
| p-value | <0.001 | <0.001 | 0.982 | 0.098 | <0.001 | |
| AOC3 | ||||||
| Correlation coefficient | 0.160 | 0.087 | −0.001 | 0.052 | ||
| p-value | <0.001 | 0.016 | 0.978 | 0.147 | ||
| AOC3 (S) | ||||||
| Correlation coefficient | −0.035 | −0.034 | 0.121 | |||
| p-value | 0.335 | 0.347 | <0.001 | |||
| MAOA | ||||||
| Correlation coefficient | 0.060 | −0.043 | ||||
| p-value | 0.095 | 0.233 | ||||
| MAOB | ||||||
| Correlation coefficient | 0.245 | |||||
| p-value | <0.001 |
| Parameter | Number of Patients/Recurrence/Death | Disease-Free Survival | Overall Survival | ||
|---|---|---|---|---|---|
| Mean Survival (95% CI) Months | p-Value | Mean Survival (95% CI) Months | p-Value | ||
| LOX | 0.413 | 0.859 | |||
| Negative | 413/16/30 | 103 (102–105) | 101 (99–103) | ||
| Positive | 357/10/25 | 105 (104–107) | 102 (100–104) | ||
| AOC3 | 0.062 | 0.050 | |||
| Negative | 178/10/19 | 103 (100–106) | 100 (97–103) | ||
| Positive | 592/16/36 | 105 (104–106) | 103 (101–104) | ||
| AOC3 (S) | 0.522 | 0.558 | |||
| Negative | 681/24/50 | 105 (103–106) | 102 (101–103) | ||
| Positive | 89/2/5 | 106 (103–108) | 104 (100–107) | ||
| MAOA | 0.484 | 0.074 | |||
| Negative | 546/20/45 | 104 (103–106) | 101 (100–103) | ||
| Positive | 224/6/10 | 106 (104–107) | 104 (102–106) | ||
| MAOB | 0.890 | 0.106 | |||
| Negative | 658/22/43 | 105 (104–106) | 103 (101–104) | ||
| Positive | 112/4/12 | 105 (102–107) | 100 (95–104) | ||
| MAOB (S) | 0.126 | 0.674 | |||
| Negative | 648/19/45 | 105 (104–106) | 102 (101–104) | ||
| Positive | 122/7/10 | 103 (99–106) | 102 (99–105) | ||
| Antibody | Company | Clone | Dilution |
|---|---|---|---|
| Amine oxidase | |||
| lysyl oxidase (LOX) | Abcam, Cambridge, UK | Polyclonal | 1:100 |
| amine oxidase (AOC3) | Abcam, Cambridge, UK | Polyclonal | 1:1000 |
| monoamine oxidase A | Abcam, Cambridge, UK | EPR7101 | 1:100 |
| monoamine oxidase B | Abcam, Cambridge, UK | Polyclonal | 1:100 |
| Molecular subtype-related proteins | |||
| ER | Thermo Scientific, San Diego, CA, USA | SP1 | 1:100 |
| PR | DAKO, Glostrup, Denmark | PgR | 1:50 |
| HER-2 | DAKO, Glostrup, Denmark | Polyclonal | 1:1500 |
| Ki-67 | Abcam, Cambridge, UK | SP6 | 1:100 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.Y.; Choi, J.; Cha, Y.J.; Koo, J.S. Evaluation of the Expression of Amine Oxidase Proteins in Breast Cancer. Int. J. Mol. Sci. 2017, 18, 2775. https://doi.org/10.3390/ijms18122775
Sun WY, Choi J, Cha YJ, Koo JS. Evaluation of the Expression of Amine Oxidase Proteins in Breast Cancer. International Journal of Molecular Sciences. 2017; 18(12):2775. https://doi.org/10.3390/ijms18122775
Chicago/Turabian StyleSun, Woo Young, Junjeong Choi, Yoon Jin Cha, and Ja Seung Koo. 2017. "Evaluation of the Expression of Amine Oxidase Proteins in Breast Cancer" International Journal of Molecular Sciences 18, no. 12: 2775. https://doi.org/10.3390/ijms18122775
APA StyleSun, W. Y., Choi, J., Cha, Y. J., & Koo, J. S. (2017). Evaluation of the Expression of Amine Oxidase Proteins in Breast Cancer. International Journal of Molecular Sciences, 18(12), 2775. https://doi.org/10.3390/ijms18122775