Identification of Autophagy-Related Genes and Their Regulatory miRNAs Associated with Celiac Disease in Children
Abstract
:1. Introduction
2. Results
2.1. Identification of Autophagy Genes/miRNAs Associated with CD
2.2. Bioinformatics Performance of the Investigated Targets as Potential CD Biomarkers
2.3. In Vitro Modulation of miR-17 Affects Autophagic Status
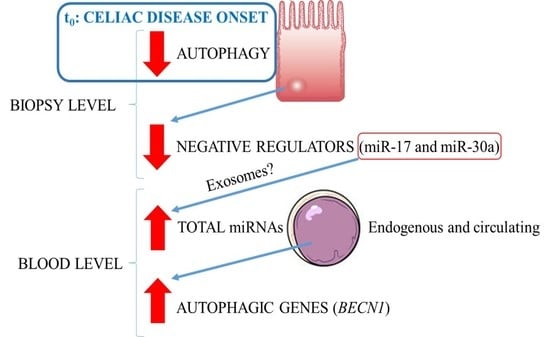
3. Discussion
4. Materials and Methods
4.1. Investigated Celiac Patients and Controls
4.2. Real-Time PCR Expression Analysis
4.3. Digested Gliadin In Vitro Assay
4.4. Immunofluorescence and Immunoblotting Analysis
4.5. Exosome Isolation, Visualization and Electroporation
4.6. Acridine Orange Staining of Autophagic Vesicles
4.7. Statistical Analysis
5. Conclusion
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Di Sabatino, A.; Corazza, G.R. Coeliac disease. Lancet 2009, 373, 1480–1493. [Google Scholar] [CrossRef]
- Bozzola, M.; Meazza, C.; Nastasio, S.; Maggiore, G. Immunology and Immune System Disorders: Celiac Disease: An Update; Nova Science Publisher: New York, NY, USA, 2014. [Google Scholar]
- Husby, S.; Koletzko, S.; Korponay-Szabó, I.R.; Mearin, M.L.; Phillips, A.; Shamir, R.; Troncone, R.; Giersiepen, K.; Branski, D.; Catassi, C.; et al. European Society for Pediatric Gastroenterology, Hepatology, and Nutrition guidelines for the diagnosis of coeliac disease. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 136–160. [Google Scholar] [CrossRef] [PubMed]
- Larrea, E.; Sole, C.; Manterola, L.; Goicoechea, I.; Armesto, M.; Arestin, M.; Caffarel, M.M.; Araujo, A.M.; Araiz, M.; Fernandez-Mercado, M.; et al. New concepts in cancer biomarkers: Circulating miRNAs in liquid biopsies. Int. J. Mol. Sci. 2016, 17, 627. [Google Scholar] [CrossRef] [PubMed]
- Kaleb, M.; Pauley, S.C.; Chan, E.K.L. MicroRNA in autoimmunity and autoimmune diseases. J. Autoimmun. 2009, 32, 189–194. [Google Scholar]
- Schaefer, J.S.; Attumi, T.; Opekun, A.R.; Abraham, B.; Hou, J.; Shelby, H.; Graham, D.Y.; Streckfus, C.; Klein, J.R. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015, 16, 5. [Google Scholar] [CrossRef] [PubMed]
- Iborra, M.; Bernuzzi, F.; Correale, C.; Vetrano, S.; Fiorino, G.; Beltrán, B.; Marabita, F.; Locati, M.; Spinelli, A.; Nos, P.; et al. Identification of serum and tissue micro-RNA expression profiles in different stages of inflammatory bowel disease. Clin. Exp. Immunol. 2013, 173, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Benderska, N.; Dittrich, A.L.; Knaup, S.; Rau, T.T.; Neufert, C.; Wach, S.; Fahlbusch, F.B.; Rauh, M.; Wirtz, R.M.; Agaimy, A.; et al. miRNA-26b Overexpression in ulcerative colitis-associated carcinogenesis. Inflamm. Bowel Dis. 2015, 21, 2039–2051. [Google Scholar] [CrossRef] [PubMed]
- Richmond, C.A.; Breault, D.T. Regulation of gene expression in the intestinal epithelium. Prog. Mol. Biol. Transl. Sci. 2010, 96, 207–229. [Google Scholar] [PubMed]
- Bascuñán-Gamboa, K.A.; Araya-Quezada, M.; Pérez-Bravo, F. MicroRNAs: An epigenetic tool to study celiac disease. Rev. Esp. Enferm. Dig. 2014, 106, 325–333. [Google Scholar] [PubMed]
- Buoli Comani, G.; Panceri, R.; Dinelli, M.; Biondi, A.; Mancuso, C.; Meneveri, R.; Barisani, D. miRNA-regulated gene expression differs in celiac disease patients according to the age of presentation. Genes Nutr. 2015, 10, 482. [Google Scholar] [CrossRef] [PubMed]
- Capuano, M.; Iaffaldano, L.; Tinto, N.; Montanaro, D.; Capobianco, V.; Izzo, V.; Tucci, F.; Troncone, G.; Greco, L.; Sacchetti, L. MicroRNA-449a overexpression, reduced NOTCH1 signals and scarce goblet cells characterize the small intestine of celiac patients. PLoS ONE 2011, 6, e29094. [Google Scholar] [CrossRef] [PubMed]
- Vaira, V.; Roncoroni, L.; Barisani, D.; Gaudioso, G.; Bosari, S.; Bulfamante, G.; Doneda, L.; Conte, D.; Tomba, C.; Bardella, M.T.; et al. microRNA profiles in coeliac patients distinguish different clinical phenotypes and are modulated by gliadin peptides in primary duodenal fibroblasts. Clin. Sci. 2014, 126, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Magni, S.; Buoli Comani, G.; Elli, L.; Vanessi, S.; Ballarini, E.; Nicolini, G.; Rusconi, M.; Castoldi, M.; Meneveri, R.; Muckenthaler, M.U.; et al. miRNAs affect the expression of innate and adaptive immunity proteins in celiac disease. Am. J. Gastroenterol. 2014, 109, 1662–1674. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Kroemer, G. SnapShot: Macroautophagy. Cell 2008, 132, 162.e1–162.e3. [Google Scholar] [CrossRef] [PubMed]
- Deretic, V.; Saitoh, T.; Akira, S. Autophagy in infection, inflammation and immunity. Nat. Rev. Immunol. 2013, 13, 722–737. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.H.; Cuervo, A.M.; Kumar, A.; Peterhoff, C.M.; Schmidt, S.D.; Lee, J.H.; Mohan, P.S.; Mercken, M.; Farmery, M.R.; et al. Macroautophagy—A novel β-amyloid peptide-generating pathway activated in Alzheimer’s disease. J. Cell Biol. 2005, 171, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Winslow, A.R.; Chen, C.W.; Corrochano, S.; Acevedo-Arozena, A.; Gordon, D.E.; Peden, A.A.; Lichtenberg, M.; Menzies, F.M.; Ravikumar, B.; Imarisio, S.; et al. α-Synuclein impairs macroautophagy: Implications for Parkinson’s disease. J. Cell Biol. 2010, 190, 1023–1037. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Vicente, M.; Talloczy, Z.; Wong, E.; Tang, G.; Koga, H.; Kaushik, S.; de Vries, R.; Arias, E.; Harris, S.; Sulzer, D.; et al. Cargo recognition failure is responsible for inefficient autophagy in Huntington’s disease. Nat. Neurosci. 2010, 13, 567–576. [Google Scholar] [CrossRef] [PubMed]
- Scharl, M.; Rogler, G. Inflammatory bowel disease: Dysfunction of autophagy? Dig. Dis. 2012, 30, 12–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henckaerts, L.; Cleynen, I.; Brinar, M.; John, J.M.; van Steen, K.; Rutgeerts, P.; Vermeire, S. Genetic variation in the autophagy gene ULK1 and risk of Crohn’s disease. Inflamm. Bowel Dis. 2011, 17, 1392–1397. [Google Scholar] [CrossRef] [PubMed]
- Pattison, J.S.; Osinska, H.; Robbins, J. Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes. Circ. Res. 2011, 109, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Aita, V.M.; Liang, X.H.; Murty, V.V.; Pincus, D.L.; Yu, W.; Cayanis, E.; Kalachikov, S.; Gilliam, T.C.; Levine, B. Cloning and genomic organization of Beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999, 59, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Kang, R.; Zeh, H.J.; Lotze, M.T.; Tang, D. The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ. 2011, 18, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef] [PubMed]
- Hong, L.; Lai, M.; Chen, M.; Xie, C.; Liao, R.; Kang, Y.J.; Xiao, C.; Hu, WY.; Han, J.; Sun, P. The miR-17–92 cluster of microRNAs confers tumorigenicity by inhibiting oncogene-induced senescence. Cancer Res. 2010, 70, 8547–8557. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, N.; Kanwar, J.R.; Kanwar, R.K.; Krishnakumar, S. Blocking the maturation of OncomiRNAs using pri-miRNA-17~92 aptamer in retinoblastoma. Nucleic Acid Ther. 2015, 25, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Dellago, H.; Bobbili, M.R.; Grillari, J. MicroRNA-17–5p: At the Crossroads of Cancer and Aging—A Mini-Review. Gerontology 2017, 63, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Comincini, S.; Allavena, G.; Palumbo, S.; Morini, M.; Durando, F.; Angeletti, F.; Pirtoli, L.; Miracco, C. microRNA-17 regulates the expression of ATG7 and modulates the autophagy process, improving the sensitivity to temozolomide and low-dose ionizing radiation treatments in human glioblastoma cells. Cancer Biol. Ther. 2013, 14, 574–586. [Google Scholar] [CrossRef] [PubMed]
- Kuma, A.; Hatano, M.; Matsui, M.; Yamamoto, A.; Nakaya, H.; Yoshimori, T.; Ohsumi, Y.; Tokuhisa, T.; Mizushima, N. The role of autophagy during the early neonatal starvation period. Nature 2004, 432, 1032–1036. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Mei, Y.; Li, K.; Huang, X.; Yang, H. Downregulation of miR-17-92a cluster promotes autophagy induction in response to celastrol treatment in prostate cancer cells. Biochem. Biophys. Res. Commun. 2016, 478, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zhang, J.; Wang, Z.; Wang, T.; Yu, Y.; He, J.; Zhang, H.; Yang, T.; Shen, Z. MicroRNA-17 regulates autophagy to promote hepatic ischemia/reperfusion injury via suppression of signal transductions and activation of transcription-3 expression. Liver Transpl. 2016, 22, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Chang, T.C.; Yu, D.; Lee, Y.S.; Wentzel, E.A.; Arking, D.E.; West, K.M.; Dang, C.V.; Thomas-Tikhonenko, A.; Mendell, J.T. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat. Genet. 2008, 40, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Kumarswamy, R.; Mudduluru, G.; Ceppi, P.; Muppala, S.; Kozlowski, M.; Niklinski, J.; Papotti, M.; Allgayer, H. MicroRNA-30a inhibits epithelial-to-mesenchymal transition by targeting Snai1 and is downregulated in non-small cell lung cancer. Int. J. Cancer 2012, 130, 2044–2053. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Tang, Q.; Qin, D.; Yu, L.; Huang, R.; Lv, G.; Zou, Z.; Jiang, X.C.; Zou, C.; Liu, W.; et al. Role of microRNA 30a targeting insulin receptor substrate 2 in colorectal tumorigenesis. Mol. Cell. Biol. 2015, 35, 988–1000. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, N.; Osera, C.; Fassina, L.; Amadio, M.; Angeletti, F.; Morini, M.; Magenes, G.; Venturini, L.; Biggiogera, M.; et al. Autophagy is modulated in human neuroblastoma cells through direct exposition to low frequency electromagnetic fields. J. Cell. Physiol. 2014, 229, 1776–1786. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Liang, J.; Li, Y.; Li, J.; Yang, X.; Zhang, X.; Han, S.; Li, S.; Li, J. Down-regulation of miRNA-30a alleviates cerebral ischemic injury through enhancing beclin 1-mediated autophagy. Neurochem. Res. 2014, 39, 1279–1291. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Liu, S.; Chen, H.; Lao, L. MicroRNA-30a downregulation contributes to chemoresistance of osteosarcoma cells through activating Beclin-1-mediated autophagy. Oncol. Rep. 2016, 35, 1757–1763. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Meng, T.; Wang, Q.S.; Jin, H.Z.; Sun, Z.Q.; Jin, B.; Fang, F.; Wang, H.J. Association of Beclin-1 and microRNA-30a expression with the severity and treatment response of colorectal cancer. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Barone, M.V.; Troncone, R.; Auricchio, S. Gliadin peptides as triggers of the proliferative and stress/innate immune response of the celiac small intestinal mucosa. Int. J. Mol. Sci. 2014, 15, 20518–20537. [Google Scholar] [CrossRef] [PubMed]
- Fassina, L.; Magenes, G.; Inzaghi, A.; Palumbo, S.; Allavena, G.; Miracco, C.; Pirtoli, L.; Biggiogera, M.; Comincini, S. AUTOCOUNTER, an ImageJ JavaScript to analyze LC3B-GFP expression dynamics in autophagy-induced astrocytoma cells. Eur. J. Histochem. 2012, 56, e44. [Google Scholar] [CrossRef] [PubMed]
- Green, P.H. The many faces of celiac disease: Clinical presentation of celiac disease in the adult population. Gastroenterology 2005, 128, S74–S78. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.M.; Ryter, S.W.; Levine, B. Autophagy in human health and disease. N. Engl. J. Med. 2013, 368, 651–662. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Levine, B. Autophagy in mammalian development and differentiation. Nat. Cell Biol. 2010, 12, 823–830. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, E.; Lippens, S.; Vanden Berghe, T.; Romagnoli, A.; Fimia, G.M.; Piacentini, M.; Vandenabeele, P. Beclin1: A role in membrane dynamics and beyond. Autophagy 2012, 8, 6–17. [Google Scholar] [CrossRef] [PubMed]
- Fu, L.L.; Cheng, Y.; Liu, B. Beclin-1: Autophagic regulator and therapeutic target in cancer. Int. J. Biochem. Cell Biol. 2013, 45, 921–924. [Google Scholar] [CrossRef] [PubMed]
- Ra, E.A.; Lee, T.A.; Won Kim, S.; Park, A.; Choi, H.J.; Jang, I.; Kang, S.; Hee Cheon, J.; Cho, J.W.; Eun Lee, J.; et al. TRIM31 promotes Atg5/Atg7-independent autophagy in intestinal cells. Nat. Commun. 2016, 7, 11726. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Arakawa, S.; Fujitani, K.; Yamaguchi, H.; Mizuta, T.; Kanaseki, T.; Komatsu, M.; Otsu, K.; Tsujimoto, Y.; Shimizu, S. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009, 461, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Wittkopf, N.; Günther, C.; Martini, E.; Waldner, M.; Amann, K.U.; Neurath, M.F.; Becker, C. Lack of intestinal epithelial atg7 affects paneth cell granule formation but does not compromise immune homeostasis in the gut. Clin. Dev. Immunol. 2012, 2012, 278059. [Google Scholar] [CrossRef] [PubMed]
- Randall-Demllo, S.; Chieppa, M.; Eri, R. Intestinal epithelium and autophagy: Partners in gut homeostasis. Front. Immunol. 2013, 4, 301. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wu, H.; Liu, X.; Li, B.; Chen, Y.; Ren, X.; Liu, C.G.; Yang, J.M. Regulation of autophagy by a beclin targeted microRNA, miR-30a, in cancer cells. Autophagy 2009, 5, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Rokach, L.; Maimon, O. Data Mining with Decision Tree; Series in Machine Perception and Artificial Intelligence; World Scientific Publishing Co. Pte. Ltd.: Singapore, 2014; Volume 69. [Google Scholar]
- Demsar, J.; Curk, T.; Erjavec, A.; Gorup, C.; Hocevar, T.; Milutinovic, M.; Mozina, M.; Polajnar, M.; Toplak, M.; Staric, A.; et al. Orange: Data Mining Toolbox in Python. J. Mach. Lear. Res. 2013, 22, 2349–2353. [Google Scholar]
- Bendifallah, S.; Daraï, E.; Ballester, M. Predictive Modeling: A New Paradigm for Managing Endometrial Cancer. Ann. Surg. Oncol. 2016, 23, 975–988. [Google Scholar] [CrossRef] [PubMed]
- Comincini, S.; Ferrara, V.; Arias, A.; Malovini, A.; Azzalin, A.; Ferretti, L.; Benericetti, E.; Cardarelli, M.; Gerosa, M.; Passarin, M.G.; et al. Diagnostic value of PRND gene expression profiles in astrocytomas: Relationship to tumor grades of malignancy. Oncol. Rep. 2007, 17, 989–996. [Google Scholar] [CrossRef] [PubMed]
- Comincini, S.; Paolillo, M.; Barbieri, G.; Palumbo, S.; Sbalchiero, E.; Azzalin, A.; Russo, M.A.; Schinelli, S. Gene expression analysis of an EGFR indirectly related pathway identified PTEN and MMP9 as reliable diagnostic markers for human glial tumor specimens. J. Biomed. Biotechnol. 2009, 2009, 924565. [Google Scholar] [CrossRef] [PubMed]
- Madsen, L.T. Cancer Prediction Nomograms for Advanced Practitioners in Oncology. J. Adv. Pract. Oncol. 2014, 5, 380–382. [Google Scholar] [PubMed]
- Lindfors, K.; Rauhavirta, T.; Stenman, S.; Mäki, M.; Kaukinen, K. In vitro models for gluten toxicity: Relevance for celiac disease pathogenesis and development of novel treatment options. Exp. Biol. Med. 2012, 237, 119–125. [Google Scholar] [CrossRef] [PubMed]
- Khaleghi, S.; Ju, J.M.; Lamba, A.; Murray, J.A. The potential utility of tight junction regulation in celiac disease: Focus on larazotide acetate. Ther. Adv. Gastroenterol. 2016, 9, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Paglin, S.; Hollister, T.; Delohery, T.; Hackett, N.; McMahill, M.; Sphicas, E.; Domingo, D.; Yahalom, J. A novel response of cancer cells to radiation involves autophagy and formation of acidic vesicles. Cancer Res. 2001, 61, 439–444. [Google Scholar] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Sbalchiero, E.; Azzalin, A.; Palumbo, S.; Barbieri, G.; Arias, A.; Simonelli, L.; Ferretti, L.; Comincini, S. Altered cellular distribution and sub-cellular sorting of doppel (Dpl) protein in human astrocytoma cell lines. Cell. Oncol. 2008, 30, 337–347. [Google Scholar] [PubMed]
- Barbieri, G.; Palumbo, S.; Gabrusiewicz, K.; Azzalin, A.; Marchesi, N.; Spedito, A.; Biggiogera, M.; Sbalchiero, E.; Mazzini, G.; Miracco, C.; et al. Silencing of cellular prion protein (PrPC) expression by DNA-antisense oligonucleotides induces autophagy-dependent cell death in glioma cells. Autophagy 2011, 7, 840–853. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, F.; Fossati, G.; Pattarozzi, A.; Würth, R.; Solari, A.; Daga, A.; Masiello, I.; Barbieri, F.; Florio, T.; Comincini, S. Inhibition of the Autophagy Pathway Synergistically Potentiates the Cytotoxic Activity of Givinostat (ITF2357) on Human Glioblastoma Cancer Stem Cells. Front. Mol. Neurosci. 2016, 9, 107. [Google Scholar] [CrossRef] [PubMed]










| Tissue | Gene/miRNA | Z-Score | p-Value |
|---|---|---|---|
| Blood | ATG7 | 2.13 | 0.2585 |
| BECN1 | 2.34 | 0.0189 * | |
| miR-17 | 1.13 | 0.2557 | |
| miR-30a | 1.51 | 1.1310 | |
| Intestinal biopsy | ATG7 | 2.41 | 0.0159 * |
| BECN1 | 2.48 | 0.0129 * | |
| miR-17 | 2.09 | 0.0365 * | |
| miR-30a | 2.16 | 0.0302 * |
| Tissue | Gene/miRNA | AUC | C.I. 95% | p-Value | Sensitivity | Specificity |
|---|---|---|---|---|---|---|
| Blood | ATG7 | 0.603 | 0.46–0.73 | 0.1723 | 78.26 | 47.06 |
| BECN1 | 0.683 | 0.54–0.79 | 0.012 * | 65.22 | 74.29 | |
| miR-17 | 0.605 | 0.46–0.73 | 0.1822 | 34.78 | 97.06 | |
| miR-30a | 0.632 | 0.49–0.75 | 0.0754 | 56.52 | 67.65 | |
| Intestinal biopsies | ATG7 | 0.697 | 0.55–0.81 | 0.007 * | 64.00 | 69.23 |
| BECN1 | 0.703 | 0.55–0.82 | 0.0068 * | 88.00 | 57.69 | |
| miR-17 | 0.671 | 0.52–0.79 | 0.0274 * | 56.00 | 84.62 | |
| miR-30a | 0.677 | 0.53–0.80 | 0.0238 * | 72.00 | 69.23 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Comincini, S.; Manai, F.; Meazza, C.; Pagani, S.; Martinelli, C.; Pasqua, N.; Pelizzo, G.; Biggiogera, M.; Bozzola, M. Identification of Autophagy-Related Genes and Their Regulatory miRNAs Associated with Celiac Disease in Children. Int. J. Mol. Sci. 2017, 18, 391. https://doi.org/10.3390/ijms18020391
Comincini S, Manai F, Meazza C, Pagani S, Martinelli C, Pasqua N, Pelizzo G, Biggiogera M, Bozzola M. Identification of Autophagy-Related Genes and Their Regulatory miRNAs Associated with Celiac Disease in Children. International Journal of Molecular Sciences. 2017; 18(2):391. https://doi.org/10.3390/ijms18020391
Chicago/Turabian StyleComincini, Sergio, Federico Manai, Cristina Meazza, Sara Pagani, Carolina Martinelli, Noemi Pasqua, Gloria Pelizzo, Marco Biggiogera, and Mauro Bozzola. 2017. "Identification of Autophagy-Related Genes and Their Regulatory miRNAs Associated with Celiac Disease in Children" International Journal of Molecular Sciences 18, no. 2: 391. https://doi.org/10.3390/ijms18020391
APA StyleComincini, S., Manai, F., Meazza, C., Pagani, S., Martinelli, C., Pasqua, N., Pelizzo, G., Biggiogera, M., & Bozzola, M. (2017). Identification of Autophagy-Related Genes and Their Regulatory miRNAs Associated with Celiac Disease in Children. International Journal of Molecular Sciences, 18(2), 391. https://doi.org/10.3390/ijms18020391