Hyper-Methylated Loci Persisting from Sessile Serrated Polyps to Serrated Cancers
Abstract
:1. Introduction
2. Results
3. Discussion
4. Patients and Methods
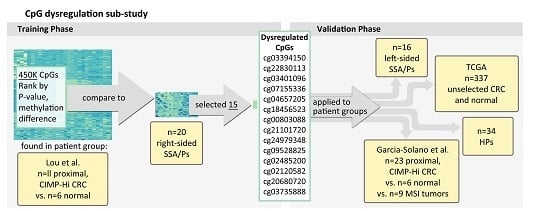
4.1. Overview
4.2. New Hampshire Colonoscopy Registry
4.3. Proximal/Right-Sided Datasets Used for Identifying Persistent Hyper-Methylated CpGs
4.4. Datasets for Assessing Hyper-Methylated CpGs across Bowel Subsites
- Left-sided SSA/P (n = 16) and HP specimens (n = 34: 55% left-sided, 45% right-sided) from the NH Colonoscopy Registry cohort.
- Colorectal cancers described in Garcia-Solano et al. GEO (GSE68060) [15] (n = 17 serrated adenocarcinomas vs. n = 11 adjacent normal mucosal samples from the Spanish cohort and n = 17 serrated adenocarcinomas vs. n = 4 adjacent normal mucosal samples from the Finnish cohort). Features used to diagnose serrated adenocarcinoma included “epithelial serrations, clear or eosinophilic cytoplasm, abundant cytoplasm, vesicular nuclei, absence of, or less than 10% necrosis of the total surface area, mucin production and cell balls and papillary rods in mucinous areas of a tumour” [17,37,38].
- Additional colorectal cancer samples that showed high microsatellite instability (MSI-H) (n = 9), as well as n = 6 normal samples from Garcia-Solano et al. [15].
- Unselected tumors from CRC patients vs. normal tissue included in the colon adenocarcinoma “COAD” dataset available from The Cancer Genome Atlas (TCGA) (n = 337) [34].
4.5. Laboratory Methods
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| SSA/P | sessile serrated adenoma/polyp |
References
- Siegel, R.; Naishadham, D.; Jemal, A. Cancer Statistics, 2013. CA Cancer J. Clin. 2013, 63, 11–30. [Google Scholar] [CrossRef] [PubMed]
- Zauber, A.G.; Winawer, S.J.; O’Brien, M.J.; Lansdorp-Vogelaar, I.; van Ballegooijen, M.; Hankey, B.F.; Shi, W.; Bond, J.H.; Schapiro, M.; Panish, J.F.; et al. Colonoscopic Polypectomy and Long-Term Prevention of Colorectal-Cancer Deaths. N. Engl. J. Med. 2012, 366, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Lieberman, D.A.; Rex, D.K.; Winawer, S.J.; Giardiello, F.M.; Johnson, D.A.; Levin, T.R. Guidelines for Colonoscopy Surveillance after Screening and Polypectomy: A Consensus Update by the Us Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012, 143, 844–857. [Google Scholar] [CrossRef] [PubMed]
- Tadros, M.; Anderson, J.C. Serrated Polyps: Clinical Implications and Future Directions. Curr. Gastroenterol. Rep. 2013, 15, 342. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.M.; Mitchell, J.M.; Sepulveda, J.L.; Sepulveda, A.R. Molecular and Histologic Considerations in the Assessment of Serrated Polyps. Arch. Pathol. Lab. Med. 2015, 139, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Bae, J.M.; Cho, N.Y.; Kang, G.H. Distinct Features between MLH1-Methylated and Unmethylated Colorectal Carcinomas with the CPG Island Methylator Phenotype: Implications in the Serrated Neoplasia Pathway. Oncotarget 2016, 7, 14095–14111. [Google Scholar] [PubMed]
- Kim, M.S.; Lee, J.; Sidransky, D. DNA Methylation Markers in Colorectal Cancer. Cancer Metastasis. Rev. 2010, 29, 181–206. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M. A.; Kouzarides, T. Cancer Epigenetics: From Mechanism to Therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef] [PubMed]
- Sakai, E.; Nakajima, A.; Kaneda, A. Accumulation of Aberrant DNA Methylation During Colorectal Cancer Development. World J. Gastroenterol. 2014, 20, 978–987. [Google Scholar] [CrossRef] [PubMed]
- Goel, A.; Boland, C.R. Epigenetics of Colorectal Cancer. Gastroenterology 2012, 143, 1442–1460. [Google Scholar] [CrossRef] [PubMed]
- Doi, A.; Park, I.H.; Wen, B.; Murakami, P.; Aryee, M.J.; Irizarry, R.; Herb, B.; Ladd-Acosta, C.; Rho, J.; Loewer, S.; et al. Differential Methylation of Tissue- and Cancer-Specific CPG Island Shores Distinguishes Human Induced Pluripotent Stem Cells, Embryonic Stem Cells and Fibroblasts. Nat. Genet. 2009, 41, 1350–1353. [Google Scholar] [CrossRef] [PubMed]
- Kanai, Y.; Hirohashi, S. Alterations of DNA Methylation Associated with Abnormalities of DNA Methyltransferases in Human Cancers During Transition from a Precancerous to a Malignant State. Carcinogenesis 2007, 28, 2434–2442. [Google Scholar] [CrossRef] [PubMed]
- Bettstetter, M.; Dechant, S.; Ruemmele, P.; Grabowski, M.; Keller, G.; Holinski-Feder, E.; Hartmann, A.; Hofstaedter, F.; Dietmaier, W. Distinction of Hereditary Nonpolyposis Colorectal Cancer and Sporadic Microsatellite-Unstable Colorectal Cancer through Quantification of Mlh1 Methylation by Real-Time Pcr. Clin. Cancer Res. 2007, 13, 3221–3228. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Tsuchiya, K.D.; Park, D.I.; Fausel, R.; Kanngurn, S.; Welcsh, P.; Dzieciatkowski, S.; Wang, J.; Grady, W.M. RET Is a Potential Tumor Suppressor Gene in Colorectal Cancer. Oncogene 2013, 32, 2037–2047. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Solano, J.; Garcia-Solano, M.E.; Torres-Moreno, D.; Carbonell, P.; Trujillo-Santos, J.; Perez-Guillermo, M.; Conesa-Zamora, P. Biomarkers for the Identification of Precursor Polyps of Colorectal Serrated Adenocarcinomas. Cell Oncol. 2016, 39, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, N.S. Serrated Pathway and Apc (Conventional)-Type Colorectal Polyps: Molecular-Morphologic Correlations, Genetic Pathways, and Implications for Classification. Am. J. Clin. Pathol. 2006, 125, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Conesa-Zamora, P.; Garcia-Solano, J.; Mdel, C.T.; Sebastian-Leon, P.; Torres-Moreno, D.; Estrada, E.; Tuomisto, A.; Wilce, J.; Makinen, M.J.; Perez-Guillermo, M.; et al. Methylome Profiling Reveals Functions and Genes Which Are Differentially Methylated in Serrated Compared to Conventional Colorectal Carcinoma. Clin. Epigenet. 2015, 7, 101. [Google Scholar] [CrossRef] [PubMed]
- Schreiner, M.A.; Weiss, D.G.; Lieberman, D.A. Proximal and Large Hyperplastic and Nondysplastic Serrated Polyps Detected by Colonoscopy Are Associated with Neoplasia. Gastroenterology 2010, 139, 1497–1502. [Google Scholar] [CrossRef] [PubMed]
- Weisenberger, D.J.; Siegmund, K.D.; Campan, M.; Young, J.; Long, T.I.; Faasse, M.A.; Kang, G.H.; Widschwendter, M.; Weener, D.; Buchanan, D.; et al. CPG Island Methylator Phenotype Underlies Sporadic Microsatellite Instability and Is Tightly Associated with Braf Mutation in Colorectal Cancer. Nat. Genet. 2006, 38, 787–793. [Google Scholar] [CrossRef] [PubMed]
- Issa, J.P. CPG Island Methylator Phenotype in Cancer. Nat. Rev. Cancer 2004, 4, 988–993. [Google Scholar] [CrossRef] [PubMed]
- Mokarram, P.; Kumar, K.; Brim, H.; Naghibalhossaini, F.; Saberi-firoozi, M.; Nouraie, M.; Green, R.; Lee, E.; Smoot, D.T.; Ashktorab, H. Distinct High-Profile Methylated Genes in Colorectal Cancer. PLoS ONE 2009, 4, e7012. [Google Scholar] [CrossRef] [PubMed]
- Draht, M.X.; Smits, K.M.; Tournier, B.; Jooste, V.; Chapusot, C.; Carvalho, B.; Cleven, A.H.; Derks, S.; Wouters, K.A.; Belt, E.J.T.; et al. Promoter Cpg Island Methylation of RET Predicts Poor Prognosis in Stage II Colorectal Cancer Patients. Mol. Oncol. 2014, 8, 679–688. [Google Scholar] [CrossRef] [PubMed]
- Bujko, M.; Kober, P.; Mikula, M.; Ligaj, M.; Ostrowski, J.; Siedlecki, J.A. Expression Changes of Cell-Cell Adhesion-Related Genes in Colorectal Tumors. Oncol. Lett. 2015, 9, 2463–2470. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Lu, Y.; Liu, H.; Wen, W.; Jia, D.; Wang, Y.; You, M. Genome-Wide Association and Fine Mapping of Genetic Loci Predisposing to Colon Carcinogenesis in Mice. Mol. Cancer Res. 2012, 10, 66–74. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.Y.; Boulden, J.; Katz, J.B.; Wang, L.; Meyer, T.J.; Soler, A.P.; Muller, A.J.; Prendergast, G.C. Bin1 Ablation Increases Susceptibility to Cancer During Aging, Particularly Lung Cancer. Cancer Res. 2007, 67, 7605–7612. [Google Scholar] [CrossRef] [PubMed]
- Hirabayashi, K.; Shiota, K.; Yagi, S. DNA Methylation Profile Dynamics of Tissue-Dependent and Differentially Methylated Regions During Mouse Brain Development. BMC Genom. 2013, 14, 82. [Google Scholar] [CrossRef] [PubMed]
- Pefani, D.E.; Dimaki, M.; Spella, M.; Karantzelis, N.; Mitsiki, E.; Kyrousi, C.; Symeonidou, I.E.; Perrakis, A.; Taraviras, S.; Lygerou, Z. Idas, a Novel Phylogenetically Conserved Geminin-Related Protein, Binds to Geminin and Is Required for Cell Cycle Progression. J. Biol. Chem. 2011, 286, 23234–23246. [Google Scholar] [CrossRef] [PubMed]
- Chien, H.C.; Wang, H.Y.; Su, Y.N.; Lai, K.Y.; Lu, L.C.; Chen, P.C.; Tsai, S.F.; Wu, C.I.; Hsieh, W.S.; Shen, C.K. Targeted Disruption in Mice of a Neural Stem Cell-Maintaining, Krab-Zn Finger-Encoding Gene That Has Rapidly Evolved in the Human Lineage. PLoS ONE 2012, 7, e47481. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Chen, B.; Pfutze, K.; Buch, S.; Steinke, V.; Holinski-Feder, E.; Stocker, S.; von Schonfels, W.; Becker, T.; Schackert, H.K.; et al. Genome-Wide Analysis Associates Familial Colorectal Cancer with Increases in Copy Number Variations and a Rare Structural Variation at 12p12.3. Carcinogenesis 2014, 35, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Habashy, H.O.; Powe, D.G.; Glaab, E.; Ball, G.; Spiteri, I.; Krasnogor, N.; Garibaldi, J.M.; Rakha, E.A.; Green, A.R.; Caldas, C.; et al. Rerg (Ras-Like, Oestrogen-Regulated, Growth-Inhibitor) Expression in Breast Cancer: A Marker of ER-Positive Luminal-Like Subtype. Breast Cancer Res. Treat. 2011, 128, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Farraye, F.A.; Mack, C.; Posnik, O.; O’Brien, M.J. Braf and Kras Mutations in Hyperplastic Polyps and Serrated Adenomas of the Colorectum: Relationship to Histology and CPG Island Methylation Status. Am. J. Surg. Pathol. 2004, 28, 1452–1459. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, M.J.; Zhao, Q.; Yang, S. Colorectal Serrated Pathway Cancers and Precursors. Histopathology 2015, 66, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Wong, C.J.; Kaz, A.M.; Dzieciatkowski, S.; Carter, K.T.; Morris, S.M.; Wang, J.; Willis, J.E.; Makar, K.W.; Ulrich, C.M.; et al. Differences in DNA Methylation Signatures Reveal Multiple Pathways of Progression from Adenoma to Colorectal Cancer. Gastroenterology 2014, 147, 418–429.e8. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas (Tcga). National Cancer Institute. Available online: http://cancergenome.nih.gov/ (accessed on 21 April 2016).
- Butterly, L.F.; Goodrich, M.; Onega, T.; Greene, M.A.; Srivastava, A.; Burt, R.; Dietrich, A. Improving the Quality of Colorectal Cancer Screening: Assessment of Familial Risk. Dig. Dis. Sci. 2010, 55, 754–760. [Google Scholar] [CrossRef] [PubMed]
- Carney, P.A.; Goodrich, M.E.; Butterly, L.F.; Dietrich, A.J. The Design and Development of a Population-Based Colonoscopy Registry. J. Registry Manag. 2006, 33, 91–99. [Google Scholar]
- Makinen, M.J. Colorectal Serrated Adenocarcinoma. Histopathology 2007, 50, 131–150. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, S.R.; Bosman, F.T.; Boffetta, P.; Ilyas, M.; Morreau, H.; Nakamura, S.I. Carcinoma of the Colon and Rectum. In Who Classification of Tumors of the Digestive System; Bosman, F.T., Carneiro, F., Hruban, R.H., Theise, N.D., Eds.; IARC: Lyon, France, 2010; pp. 134–146. [Google Scholar]
- Koestler, D.C.; Li, J.; Baron, J.A.; Tsongalis, G.J.; Butterly, L.F.; Goodrich, M.; Lesseur, C.; Karagas, M.R.; Marsit, C.J.; Moore, J.H.; et al. Distinct Patterns of DNA Methylation in Conventional Adenomas Involving the Right and Left Colon. Mod. Pathol. 2014, 27, 145–155. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.J.; Butcher, L.M.; Feber, A.; Teschendorff, A.E.; Chakravarthy, A.R.; Wojdacz, T.K.; Beck, S. Champ: 450 k Chip Analysis Methylation Pipeline. Bioinformatics 2014, 30, 428–430. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Marabita, F.; Lechner, M.; Bartlett, T.; Tegner, J.; Gomez-Cabrero, D.; Beck, S. A β-Mixture Quantile Normalization Method for Correcting Probe Design Bias in Illumina Infinium 450 k DNA Methylation Data. Bioinformatics 2013, 29, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Storey, J.D. Capturing Heterogeneity in Gene Expression Studies by Surrogate Variable Analysis. PLoS Genet. 2007, 3, 1724–1735. [Google Scholar] [CrossRef] [PubMed]
- Teschendorff, A.E.; Zhuang, J.; Widschwendter, M. Independent Surrogate Variable Analysis to Deconvolve Confounding Factors in Large-Scale Microarray Profiling Studies. Bioinformatics 2011, 27, 1496–1505. [Google Scholar] [CrossRef] [PubMed]
- Johnson, W.E.; Li, C.; Rabinovic, A. Adjusting Batch Effects in Microarray Expression Data Using Empirical Bayes Methods. Biostatistics 2007, 8, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Roessler, J.; Ammerpohl, O.; Gutwein, J.; Hasemeier, B.; Anwar, S.L.; Kreipe, H.; Lehmann, U. Quantitative Cross-Validation and Content Analysis of the 450k DNA Methylation Array from Illumina, Inc. BMC Res. 2012, 5, 210. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Linear Models and Empirical Bayes Methods for Assessing Differential Expression in Microarray Experiments. Stat. Appl. Genet. Mol. Biol. 2004, 3, 3. [Google Scholar] [CrossRef] [PubMed]
- Butcher, L.M.; Beck, S. Probe Lasso: A Novel Method to Rope in Differentially Methylated Regions with 450 k DNA Methylation Data. Methods 2015, 72, 21–28. [Google Scholar] [CrossRef] [PubMed]



| Characteristics | Overall Population | Methylation Study | ||
|---|---|---|---|---|
| SSA/P | SSA/P | |||
| Gender | N | % | N | % |
| Female | 38 | 58 | 12 | 60 |
| Male | 27 | 42 | 8 | 40 |
| Total | 65 | 100 | 20 | 100 |
| Age: mean, SD | 56 ± 7.8 | 57 ± 6.9 | ||
| Polyp size | ||||
| <5 mm | 26 | 39 | 11 | 55 |
| 5–9 mm | 17 | 26 | 6 | 30 |
| 10–20 mm | 20 | 30 | 3 | 15 |
| CpG Site | Gene | Feature | Serr. Normal | Serr. Tumor | logFC | p-Value | BH-Corrected | Gene Name |
|---|---|---|---|---|---|---|---|---|
| Mean | Mean | p-Value | ||||||
| cg00803088 | RET | Body-island | 0.13 | 0.60 | 0.46 | 7.35 × 10−12 | 5.72 × 10−10 | RET proto-oncogene |
| cg02120582 | PDGFD | 5′UTR-island | 0.089 | 0.55 | 0.46 | 1.16 × 10−11 | 8.38 × 10−10 | Platelet derived growth factor D |
| cg02485200 | JAM2 | 1stExon-island | 0.094 | 0.55 | 0.46 | 1.13 × 10−9 | 4.08 × 10−8 | Junctional adhesion molecule 2 |
| cg03394150 | GSG1L | Body-island | 0.094 | 0.71 | 0.61 | 1.91 × 10−16 | 1.77 × 10−13 | GSG1-like |
| cg03401096 | MIR4493 | IGR-island | 0.086 | 0.63 | 0.54 | 3.19 × 10−13 | 4.41 × 10−11 | microRNA-4493 |
| cg03735888 | ZNF132 | TSS200-island | 0.061 | 0.61 | 0.55 | 4.10 × 10−14 | 8.61 × 10−12 | Zinc finger protein 132 |
| cg04657205 | MIR1233-1 | IGR-island | 0.094 | 0.52 | 0.42 | 5.44 × 10−6 | 6.21 × 10−5 | microRNA-1233-1 |
| cg07155336 | NTNG1 | 5′UTR-island | 0.075 | 0.64 | 0.56 | 1.18 × 10−10 | 5.91 × 10−9 | Netrin G1 |
| cg09528825 | GSG1L | Body-island | 0.10 | 0.64 | 0.54 | 3.54 × 10−17 | 5.57 × 10−14 | GSG1-like |
| cg14553600 | JAM2 | 1stExon-island | 0.12 | 0.53 | 0.41 | 2.27 × 10−10 | 1.03 × 10−8 | Junctional adhesion molecule 2 |
| cg18456523 | MCIDAS | IGR-island | 0.077 | 0.52 | 0.44 | 9.16 × 10−6 | 9.69 × 10−5 | Multiciliate differentiation, DNA synthesis |
| cg20680720 | ZNF568 | TSS200-island | 0.051 | 0.58 | 0.53 | 5.44 × 10−11 | 3.08 × 10−9 | Zinc finger protein 568 |
| cg21101720 | ANKRD13B | Body-island | 0.13 | 0.68 | 0.55 | 2.71 × 10−11 | 1.71 × 10-−9 | Ankyrin repeat domain 13B |
| cg22830113 | BIN1 | IGR-island | 0.078 | 0.56 | 0.48 | 1.61 × 10−7 | 2.95 × 10−6 | Bridging integrator 1 |
| cg24979348 | RERG | 5′UTR-island | 0.091 | 0.52 | 0.43 | 8.02 × 10−8 | 1.61 × 10−6 | RAS-like, estrogen-regulated, growth inhib |
| Gene | Differentially Methylated Region | Serr. Normal | Serr. Tumor | logFC | BH-Corrected | |||
|---|---|---|---|---|---|---|---|---|
| Start | End | Size | Core Size | Mean | Mean | DMR p-Value | ||
| GSG1L | 28074304 | 28075323 | 1020 | 912 | 0.12 | 0.63 | 0.51 | 1.46 × 10−91 |
| MIR4493 | 123228869 | 123229236 | 368 | 149 | 0.10 | 0.53 | 0.42 | 3.04 × 10−26 |
| NTNG1 | 108023286 | 108023663 | 378 | 117 | 0.43 | 0.66 | 0.23 | 2.18 × 10−19 |
| MCIDAS | 54518582 | 54519046 | 465 | 357 | 0.25 | 0.64 | 0.39 | 1.57 × 10−35 |
| ZNF568 | 37407207 | 37407307 | 101 | 69 | 0.10 | 0.59 | 0.49 | 7.35 × 10−28 |
| RERG | 15374496 | 15374673 | 178 | 50 | 0.07 | 0.42 | 0.35 | 3.43 × 10-11 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Andrew, A.S.; Baron, J.A.; Butterly, L.F.; Suriawinata, A.A.; Tsongalis, G.J.; Robinson, C.M.; Amos, C.I. Hyper-Methylated Loci Persisting from Sessile Serrated Polyps to Serrated Cancers. Int. J. Mol. Sci. 2017, 18, 535. https://doi.org/10.3390/ijms18030535
Andrew AS, Baron JA, Butterly LF, Suriawinata AA, Tsongalis GJ, Robinson CM, Amos CI. Hyper-Methylated Loci Persisting from Sessile Serrated Polyps to Serrated Cancers. International Journal of Molecular Sciences. 2017; 18(3):535. https://doi.org/10.3390/ijms18030535
Chicago/Turabian StyleAndrew, Angeline S., John A. Baron, Lynn F. Butterly, Arief A. Suriawinata, Gregory J. Tsongalis, Christina M. Robinson, and Christopher I. Amos. 2017. "Hyper-Methylated Loci Persisting from Sessile Serrated Polyps to Serrated Cancers" International Journal of Molecular Sciences 18, no. 3: 535. https://doi.org/10.3390/ijms18030535
APA StyleAndrew, A. S., Baron, J. A., Butterly, L. F., Suriawinata, A. A., Tsongalis, G. J., Robinson, C. M., & Amos, C. I. (2017). Hyper-Methylated Loci Persisting from Sessile Serrated Polyps to Serrated Cancers. International Journal of Molecular Sciences, 18(3), 535. https://doi.org/10.3390/ijms18030535