Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium bifidum
Abstract
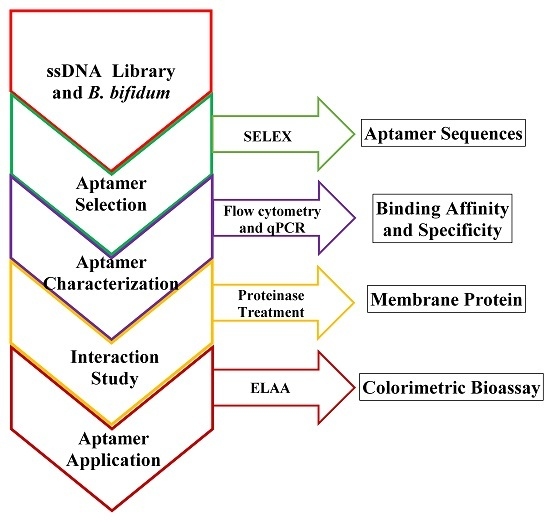
:1. Introduction
2. Results and Discussion
2.1. SELEX Optimization
2.2. Determination of Affinity and Specificity
2.3. Aptamer Truncations and Their Effects on the Binding Ability to B. bifidum
2.4. Proteinase Treatment for Bacteria
2.5. Colorimetric Detection of B. bifidum
3. Materials and Methods
3.1. Reagents and Apparatus
3.2. DNA Library and PCR Amplification
3.3. Aptamer Selection
3.4. Flow-Cytometric Analysis
3.5. Aptamer CCFM641-5 Binding Assays by Quantitative PCR (qPCR)
3.6. Aptamer Truncation
3.7. Proteinase Treatment for Bacteria
3.8. Colorimetric Bioassay on the Basis of the Selected Aptamer
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yu, Y.; Jiang, F.; Zhou, J.; Li, Y.; Liang, C.; Dang, L.; Lu, A.; Zhang, G. Development of Cell-SELEX technology and its application in cancer diagnosis and therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef] [PubMed]
- Famulok, M.; Hartig, J.S.; Mayer, G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007, 107, 3715–3743. [Google Scholar] [CrossRef] [PubMed]
- Navani, N.K.; Li, Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. 2006, 10, 272–281. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, H.P.; Smiley, R.D.; Jaykus, L.A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 2010, 87, 2323–2334. [Google Scholar] [CrossRef] [PubMed]
- Marton, S.; Cleto, F.; Krieger, M.A.; Cardoso, J. Isolation of an aptamer that binds specifically to E. coli. PLoS ONE 2016, 11, e0153637. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.A.; Zhang, H.; Guan, L.L.; Li, X.F.; Le, X.C. Selection of aptamers against live bacterial cells. Anal. Chem. 2008, 80, 7812–7819. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zhou, J.; Luo, F.L.; Mohammed, A.B.; Zhang, X.L. Aptamer from whole-bacterium SELEX as new therapeutic reagent against virulent Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2007, 357, 743–748. [Google Scholar] [CrossRef] [PubMed]
- Duan, N.; Wu, S.; Chen, X.; Huang, Y.; Wang, Z. Selection and identification of a DNA aptamer targeted to Vibrio parahemolyticus. J. Agric. Food Chem. 2012, 60, 4034–4038. [Google Scholar] [CrossRef] [PubMed]
- Hamula, C.L.A.; Le, X.C.; Li, X.F. DNA aptamers binding to multiple prevalent M-types of Streptococcus pyogenes. Anal. Chem. 2011, 83, 3640–3647. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Li, S.; Chen, L.; Ding, H.; Xu, H.; Huang, Y.; Li, J.; Liu, N.; Cao, W.; Zhu, Y.; Shen, B.; Shao, N. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009, 37, 4621–4628. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kunisawa, J.; Kweon, M.N.; Ji, G.E.; Kiyono, H. Oral feeding of Bifidobacterium bifidum (BGN4) prevents CD4+ CD45RBhigh T cell-mediated inflammatory bowel disease by inhibition of disordered T cell activation. Clin. Immunol. 2007, 123, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Repa, A.; Thanhaeuser, M.; Endress, D.; Weber, M.; Kreissl, A.; Binder, C.; Berger, A.; Haiden, N. Probiotics (Lactobacillus acidophilus and Bifidobacterium bifidum) prevent NEC in VLBW infants fed breast milk but not formula. Pediatr. Res. 2015, 77, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, I.; Turroni, F.; Piemontese, A.; Mancabelli, L.; Milani, C.; Viappiani, A.; Prevedini, G.; Sanchez, B.; Margolles, A.; Elviri, L.; et al. Evidence for cholesterol-lowering activity by Bifidobacterium bifidum PRL2010 through gut microbiota modulation. Appl. Microbiol. Biotechnol. 2015, 99, 6813–6829. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.J.; Qiu, L.H.; Guan, R.Z.; Hu, S.J.; Liu, Y.Y.; Wang, G.J. Protective effect of probiotics in the treatment of infantile eczema. Exp. Ther. Med. 2015, 9, 1593–1596. [Google Scholar] [CrossRef] [PubMed]
- Turroni, F.; Taverniti, V.; Ruas-Madiedo, P.; Duranti, S.; Guglielmetti, S.; Lugli, G.A.; Gioiosa, L.; Palanza, P.; Margolles, A.; van Sinderen, D.; et al. Bifidobacterium bifidum PRL2010 modulates the host innate immune response. Appl. Environ. Microbiol. 2014, 80, 730–740. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, J.M.; Bauman, N.A.; Oung, I.; Perman, J.A.; Yolken, R.H. Feeding of Bifidobacterium bifidum and Streptococcus thermophilus to infants in hospital for prevention of diarrhoea and shedding of rotavirus. Lancet 1994, 344, 1046–1049. [Google Scholar] [CrossRef]
- López, P.; González-Rodríguez, I.; Gueimonde, M.; Margolles, A.; Suárez, A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS ONE 2011, 6, e24776. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, J.; Britz, T.J.; Torriani, S.; Witthuhn, R.C. Identification of probiotic microorganisms in South African products using PCR-based DGGE analysis. Int. J. Food Microbiol. 2005, 98, 11–21. [Google Scholar] [CrossRef] [PubMed]
- Mazzola, G.; Aloisio, I.; Biavati, B.; Di Gioia, D. Development of a synbiotic product for newborns and infants. LWT-Food Sci. Technol. 2015, 64, 727–734. [Google Scholar] [CrossRef]
- Stanton, C.; Gardiner, G.; Meehan, H.; Collins, K.; Fitzgerald, G.; Lynch, P.B.; Ross, R.P. Market potential for probiotics. Am. J. Clin. Nutr. 2001, 73, 476–483. [Google Scholar]
- Dinoto, A.; Marques, T.M.; Sakamoto, K.; Fukiya, S.; Watanabe, J.; Ito, S.; Yokota, A. Population dynamics of Bifidobacterium species in human feces during raffinose administration monitored by fluorescence in situ hybridization-flow cytometry. Appl. Environ. Microbiol. 2006, 72, 7739–7747. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Fujimoto, J.; Kado, Y.; Takada, T.; Matsumoto, K.; Tanaka, R. Quantitative PCR with 16S rRNA-gene-targeted species-specific primers for analysis of human intestinal bifidobacteria. Appl. Environ. Microbiol. 2004, 70, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Mullié, C.; Odou, M.F.; Singer, E.; Romond, M.B.; Izard, D. Multiplex PCR using 16S rRNA gene-targeted primers for the identifcation of bifidobacteria from human origin. FEMS Microbiol. Lett. 2003, 222, 129–136. [Google Scholar] [CrossRef]
- Vincent, D.; Roy, D.; Mondou, F.; Déry, C. Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 1998, 43, 185–193. [Google Scholar] [CrossRef]
- Torres-Chavolla, E.; Alocilja, E.C. Aptasensors for detection of microbial and viral pathogens. Biosens. Bioelectron. 2009, 24, 3175–3182. [Google Scholar] [CrossRef] [PubMed]
- Langendijk, P.S.; Schut, F.; Jansen, G.J.; Raangs, G.C.; Kamphuis, G.R.; Wilkinson, M.H.; Welling, G.W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 1995, 61, 3069–3075. [Google Scholar] [PubMed]
- Ikebukuro, K.; Kiyohara, C.; Sode, K. Novel electrochemical sensor system for protein using the aptamers in sandwich manner. Biosens. Bioelectron. 2005, 20, 2168–2172. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, Y.; Li, H.; Wen, Y.; Fan, X.; Lin, F.; Tan, L.; Yao, S. Ultrasensitive electrochemical aptasensor for thrombin based on the amplification of aptamer–AuNPs–HRP conjugates. Biosens. Bioelectron. 2011, 26, 2297–2303. [Google Scholar] [CrossRef] [PubMed]
- Aimaiti, R.; Qin, L.; Cao, T.; Yang, H.; Wang, J.; Lu, J.; Huang, X.; Hu, Z. Identification and application of ssDNA aptamers against H37Rv in the detection of Mycobacterium tuberculosis. Appl. Microbiol. Biotechnol. 2015, 99, 9073–9083. [Google Scholar] [CrossRef] [PubMed]
- Barthelmebs, L.; Jonca, J.; Hayat, A.; Prieto-Simon, B.; Marty, J.L. Enzyme-Linked Aptamer Assays (ELAAs), based on a competition format for a rapid and sensitive detection of Ochratoxin A in wine. Food Control 2011, 22, 737–743. [Google Scholar] [CrossRef]
- Nie, J.; Deng, Y.; Deng, Q.P.; Zhang, D.W.; Zhou, Y.L.; Zhang, X.X. A self-assemble aptamer fragment/target complex based high-throughput colorimetric aptasensor using enzyme linked aptamer assay. Talanta 2013, 106, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Cho, Y.S.; Kang, S.; Lee, E.J.; Lee, G.H.; Hah, S.S. A colorimetric sandwich-type assay for sensitive thrombin detection based on enzyme-linked aptamer assay. Anal. Biochem. 2014, 462, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Jayasena, S.D. Aptamers: An emerging class of molecules that rival antibodies in diagnostics. Clin. Chem. 1999, 45, 1628–1650. [Google Scholar] [PubMed]
- Liu, G.; Yu, X.; Xue, F.; Chen, W.; Ye, Y.; Yang, X.; Lian, Y.; Yan, Y.; Zong, K. Screening and preliminary application of a DNA aptamer for rapid detection of Salmonella O8. Microchim. Acta 2012, 178, 237–244. [Google Scholar] [CrossRef]
- Reuter, J.S.; Mathews, D.H. RNAstructure: Software for RNA secondary structure prediction and analysis. BMC Bioinform. 2010, 11, 129. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Tang, Z.; Mallikaratchy, P.; Xiao, Z.; Tan, W. Optimization and modifications of aptamers selected from live cancer cell lines. ChemBioChem 2007, 8, 603–606. [Google Scholar] [CrossRef] [PubMed]
- Stoltenburg, R.; Schubert, T.; Strehlitz, B. In vitro selection and interaction studies of a DNA aptamer targeting protein A. PLoS ONE 2015, 10, e0134403. [Google Scholar] [CrossRef] [PubMed]
- Shangguan, D.; Li, Y.; Tang, Z.; Cao, Z.C.; Chen, H.W.; Mallikaratchy, P.; Sefah, K.; Yang, C.J.; Tan, W. Aptamers evolved from live cells as effective molecular probes for cancer study. Proc. Natl. Acad. Sci. USA 2006, 103, 11838–11843. [Google Scholar] [CrossRef] [PubMed]







| Name | Sequence (5′ → 3′) |
|---|---|
| CCFM641-2 | GCCTGGCCAGGTGCCCCGATATAGCGACGCCTTGCCCGGC |
| CCFM641-4 | GCCCCGGACGGCGGGAAGCCTCGTACCCCCCGTGAGCGGC |
| CCFM641-5 | TGCGTGAGCGGTAGCCCCGTACGACCCACTGTGGTTGGGC |
| CCFM641-12 | GTCACACCGGCCGTCTCCGGTGTGGGACGCCCGCTGTGGC |
| Name | Sequence (5′ → 3′) |
|---|---|
| CCFM641-5 | AGCAGCACAGAGGTCAGATGTGCGTGAGCGGTAGCCCCGTACGACCCACTGTGGTTGGGCCCTATGCGTGCTACCGTGAA |
| CCFM641-5F | TGCGTGAGCGGTAGCCCCGTACGACCCACTGTGGTTGGGC CCTATGCGTGCTACCGTGAA |
| CCFM641-5R | AGCAGCACAGAGGTCAGATGTGCGTGAGCGGTAGCCCCGTACGACCCACTGTGGTTGGGC |
| CCFM641-5FR | TGCGTGAGCGGTAGCCCCGTACGACCCACTGTGGTTGGGC |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, L.; Wang, L.; Lu, W.; Zhao, J.; Zhang, H.; Chen, W. Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium bifidum. Int. J. Mol. Sci. 2017, 18, 883. https://doi.org/10.3390/ijms18050883
Hu L, Wang L, Lu W, Zhao J, Zhang H, Chen W. Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium bifidum. International Journal of Molecular Sciences. 2017; 18(5):883. https://doi.org/10.3390/ijms18050883
Chicago/Turabian StyleHu, Lujun, Linlin Wang, Wenwei Lu, Jianxin Zhao, Hao Zhang, and Wei Chen. 2017. "Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium bifidum" International Journal of Molecular Sciences 18, no. 5: 883. https://doi.org/10.3390/ijms18050883
APA StyleHu, L., Wang, L., Lu, W., Zhao, J., Zhang, H., & Chen, W. (2017). Selection, Characterization and Interaction Studies of a DNA Aptamer for the Detection of Bifidobacterium bifidum. International Journal of Molecular Sciences, 18(5), 883. https://doi.org/10.3390/ijms18050883