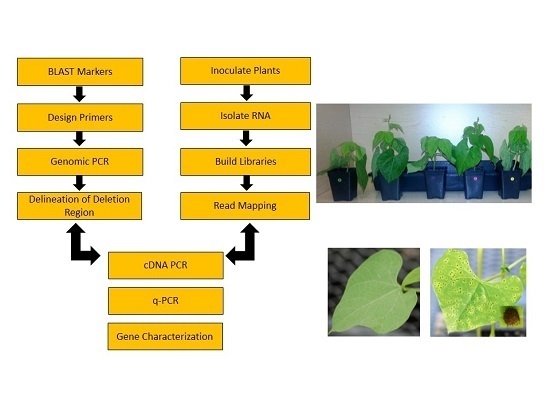
Marker-Assisted Molecular Profiling, Deletion Mutant Analysis, and RNA-Seq Reveal a Disease Resistance Cluster Associated with Uromyces appendiculatus Infection in Common Bean Phaseolus vulgaris L.
Abstract
:1. Introduction
2. Results
2.1. Marker Assisted Molecular Profiling of Genomic DNA
2.2. Illumina Sequence Data
2.3. Gene Expression during U. appendiculatus Infection
2.4. RT-PCR and Confirmation of Gene Expression
2.5. Real Time/Quantitative PCR (q-PCR)
3. Discussion
3.1. Identification of SB1 Location in Gene Phvul.010G025000
3.2. Delineation of crg Deletion Region
3.3. Current Perspective of Disease Resistance and Relation to the Bean-Bean-Rust Interaction
4. Materials and Methods
4.1. DNA Isolation
4.2. DNA Quantification and Marker Analysis
4.3. Maintenance of Pathogen and Inoculum Preparation
4.4. Plant Inoculation
4.5. RNA Isolation
4.6. cDNA Synthesis
4.7. Illumina Library Prep
4.8. Data Analysis
4.9. Real Time/Quantitative PCR (q-PCR)
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Buruchara, R.; Chirwa, R.; Sperling, L.; Mukankusi, C.; Rubyogo, J.C.; Mutonhi, R.; Abang, M. Development and delivery of bean varieties in africa: The pan-africa bean research alliance (pabra) model. Afr. Crop Sci. J. 2011, 19, 227–245. [Google Scholar]
- Petry, N.; Boy, E.; Wirth, J.P.; Hurrell, R.F. Review: The potential of the common bean (Phaseolus vulgaris) as a vehicle for iron biofortification. Nutrients 2015, 7, 1144–1173. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P. Nothing common abouth these beans. Food Blog: Food News from the UC Division of Agriculture and Natural Resources. Available online: http://ucanr.edu/blogs/blogcore/postdetail.cfm?postnum=14233 (accessed on 18 May 2017).
- McClean, P.; Cannon, S.; Gepts, P.; Hudson, M.; Jackson, S.; Rokhsar, D.; Schmutz, J.; Vance, C. Towards a Whole Genome Sequence of Common Bean, (Phaseolus vulgaris): Background, Approaches, Applications. 2008. Available online: www.researchgate.net/publication/242515421_Executive_Summary_Towards_a_Whole_Genome_Sequence_of_Common_Bean_Phaseolus_vulgaris_Background_Approaches_Applications (accessed on 19 May 2017).
- Chandalia, M.; Garg, A.; Lutjohann, D.; von Bergmann, K.; Grundy, S.M.; Brinkley, L.J. Beneficial effects of high dietary fiber intake in patients with type 2 diabetes mellitus. N. Engl. J. Med. 2000, 342, 1392–1398. [Google Scholar] [CrossRef] [PubMed]
- Namugwanya, M.; Tenywa, J.S.; Otabbong, E.; Mubiru, D.N.; Masamba, T.A. Development of common bean (Phaseolus vulgaris L.) production under low soil phosphorus and drought in sub-saharan africa: A review. J. Sustain. Dev. 2014, 7, 128. [Google Scholar] [CrossRef]
- Mamidi, S.; Rossi, M.; Moghaddam, S.M.; Annam, D.; Lee, R.; Papa, R.; McClean, P.E. Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity 2013, 110, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Schmutz, J.; McClean, P.E.; Mamidi, S.; Wu, G.A.; Cannon, S.B.; Grimwood, J.; Jenkins, J.; Shu, S.; Song, Q.; Chavarro, C.; et al. A reference genome for common bean and genome-wide analysis of dual domestications. Nat. Genet. 2014, 46, 707–713. [Google Scholar] [CrossRef] [PubMed]
- Kelly, J.D.; Copeland, J.O. Sierra. A new pinto bean for Michigan. Ext. Bull. 1994, 994, E-2494. [Google Scholar]
- Kalavacharla, V.; Stavely, J.; Myers, J.; McClean, P. Crg, a gene required for Ur-3-mediated rust resistance in common bean, maps to a resistance gene analog cluster. Mol. Plant Microbe Interact. 2000, 13, 1237–1242. [Google Scholar] [CrossRef] [PubMed]
- Dangl, J.; Jones, J. Plant pathogens and integrated defence responses to infection. Nature 2001, 411, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Coyne, C.B.; Zeh, H.J.; Lotze, M.T. PAMPS and DAMPS: Signal 0 s that spur autophagy and immunity. Immunol. Rev. 2012, 249, 158–175. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Liu, Y.; Wang, J.; He, M.; Zhou, X.; Yang, C.; Yuan, C.; Wang, J.; Chern, M.; Yin, J.; et al. The durably resistant rice cultivar digu activates defence gene expression before the full maturation of magnaporthe oryzae appressorium. Mol. Plant Pathol. 2016, 17, 354–368. [Google Scholar] [CrossRef] [PubMed]
- Block, A.; Alfano, J.R. Plant targets for pseudomonas syringae type III effectors: Virulence targets or guarded decoys? Curr. Opin. Microbiol. 2011, 14, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.T.; Dangl, J.L. Paired plant immune receptors. Science 2014, 344, 267–268. [Google Scholar] [CrossRef] [PubMed]
- Ballantyne, B. The Genetic Bases of Resistance to Rust, Caused by Uromyces Appendiculatus in Bean (Phaseolus vulgaris); University of Sydney: Sydney, Australia, 1978. [Google Scholar]
- Miklas, P.; Pastor-Corrales, M.; Jung, G.; Coyne, D.; Kelly, J.; McClean, P.; Gepts, P. Comprehensive linkage map of bean rust resistance genes. Annu. Rep. Bean Improv. Coop. 2002, 45, 125–129. [Google Scholar]
- Rivkin, M.I.; Vallejos, C.E.; McClean, P.E. Disease-resistance related sequences in common bean. Genome 1999, 42, 1–7. [Google Scholar] [CrossRef]
- Kalavacharla, V.; Liu, Z.; Meyers, B.; Thimmapuram, J.; Melmaiee, K. Identification and analysis of common bean (Phaseolus vulgaris L.) transcriptomes by massively parallel pyrosequencing. BMC Plant Biol. 2011, 11, 135. [Google Scholar] [CrossRef] [PubMed]
- Phytozome. Available online: http://phytozome.jgi.doe.gov/ (accessed on 18 May 2017).
- Melmaiee, K.; Todd, A.; McClean, P.; Lee, R.; Schlueter, J.; Jackson, S.; Kalavacharla, V. Identification of molecular markers associated with the deleted region in common bean (Phaseolus vulgaris) Ur-3 mutants. Aust. J. Crop Sci. 2013, 7, 354. [Google Scholar]
- Rupe, J.; Sconyers, L. Soybean rust. The plant health instructor. Am. Phytopathol. Soc. 2008. [Google Scholar] [CrossRef]
- Van der Biezen, E.A.; Jones, J.D. The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 1998, 8, R226–R228. [Google Scholar] [CrossRef]
- Eitas, T.; Nimchuk, Z.; Dangl, J. Arabidopsis tao1 is a TIR-NB-LRR protein that contributes to disease resistance induced by the pseudomonas syringae effector Avrb. Proc. Natl. Acad. Sci. USA 2008, 105, 6475–6480. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.B.; Brommonschenkel, S.; Chunwongse, J.; Frary, A.; Ganal, M.W.; Spivey, R.; Wu, T.; Earle, E.D.; Tanksley, S.D. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 1993, 262, 1432–1436. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, G.; Mayr, G.; Kasiem, M.M.A.; Albrecht, M.; Cornelissen, B.J.C.; Takken, F.L.W. Structure–function analysis of the NB-ARC domain of plant disease resistance proteins. J. Exp. Bot. 2008, 59, 1383–1397. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.; Godiard, L.; Straube, E.; Ashfield, T.; Lewald, J.; Sattler, A.; Innes, R.; Dangl, J. Structure of the arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 1995, 269, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Bent, A.; Kunkel, B.; Dahlbeck, D.; Brown, K.; Schmidt, R.; Giraudat, J.; Leung, J.; Staskawicz, B. RPS2 of arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 1994, 265, 1856–1860. [Google Scholar] [CrossRef] [PubMed]
- Mindrinos, M.; Katagiri, F.; Yu, G.; Ausubel, F. The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 1994, 78, 1089–1099. [Google Scholar] [CrossRef]
- Parker, J.E.; Coleman, M.J.; Szabo, V.; Frost, L.N.; Schmidt, R.; van der Biezen, E.A.; Moores, T.; Dean, C.; Daniels, M.J.; Jones, J.D.G. The arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with N and L6. Plant Cell 1997, 9, 879–894. [Google Scholar] [CrossRef] [PubMed]
- Whitman, S.; McCormick, S.; Baker, B. The n gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 8776–8781. [Google Scholar]
- Lawrence, G.J.; Finnegan, E.J.; Ayliffe, M.A.; Ellis, J.G. The L6 gene for flax rust resistance is related to the arabidopsis bacterial resistance gene RPS2 and the tobacco viral resistance gene N. Plant Cell 1995, 7, 1195–1206. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 18 May 2017).
- Gururani, M.A.; Venkatesh, J.; Upadhyaya, C.P.; Nookaraju, A.; Pandey, S.K.; Park, S.W. Plant disease resistance genes: Current status and future directions. Physiol. Mol. Plant Pathol. 2012, 78, 51–65. [Google Scholar] [CrossRef]
- Nishimura, M.T.; Monteiro, F.; Dangl, J.L. Treasure your exceptions: Unusual domains in immune receptors reveal host virulence targets. Cell 2015, 161, 957–960. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, C.; Huet, G.; Jauneau, A.; Camborde, L.; Trémousaygue, D.; Kraut, A.; Zhou, B.; Levaillant, M.; Adachi, H.; Yoshioka, H.; et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell 2015, 161, 1074–1088. [Google Scholar] [CrossRef] [PubMed]
- Sarris Panagiotis, F.; Duxbury, Z.; Huh Sung, U.; Ma, Y.; Segonzac, C.; Sklenar, J.; Derbyshire, P.; Cevik, V.; Rallapalli, G.; Saucet, S.B.; et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell 2015, 161, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Vleeshouwers, V.G.A.A.; Oliver, R.P. Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 2014, 27, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Link, T.I.; Lang, P.; Scheffler, B.E.; Duke, M.V.; Graham, M.A.; Cooper, B.; Tucker, M.L.; van de Mortel, M.; Voegele, R.T.; Mendgen, K.; et al. The haustorial transcriptomes of uromyces appendiculatus and phakopsora pachyrhizi and their candidate effector families. Mol. Plant Pathol. 2014, 15, 379–393. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.I.B.; Armstrong, M.R.; Gilroy, E.M.; Boevink, P.C.; Hein, I.; Taylor, R.M.; Zhendong, T.; Engelhardt, S.; Vetukuri, R.R.; Harrower, B.; et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 2010, 107, 9909–9914. [Google Scholar] [CrossRef] [PubMed]
- Engelhardt, S.; Boevink, P.C.; Armstrong, M.R.; Ramos, M.B.; Hein, I.; Birch, P.R. Relocalization of late blight resistance protein R3a to endosomal compartments is associated with effector recognition and required for the immune response. Plant Cell 2012, 24, 5142–5158. [Google Scholar] [CrossRef] [PubMed]
- Rafiqi, M.; Ellis, J.G.; Ludowici, V.A.; Hardham, A.R.; Dodds, P.N. Challenges and progress towards understanding the role of effectors in plant–fungal interactions. Curr. Opin. Plant Biol. 2012, 15, 477–482. [Google Scholar] [CrossRef] [PubMed]
- Hurtado-Gonzales, O.P.; Valentini, G.; Gilio, T.A.S.; Martins, A.M.; Song, Q.; Pastor-Corrales, M.A. Fine mapping of Ur-3, a historically important rust resistance locus in common bean. G3: Genes Genomes Genet. 2016, 7, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Hinrichs, A.S.; Karolchik, D.; Baertsch, R.; Barber, G.P.; Bejerano, G.; Clawson, H.; Diekhans, M.; Furey, T.S.; Harte, R.A.; Hsu, F. The UCSC genome browser database: Update 2006. Nucleic Acids Res. 2006, 34, D590–D598. [Google Scholar] [CrossRef] [PubMed]
- Doyle, J. DNA protocols for plants. In Molecular Techniques in Taxonomy; Springer: Berlin/Heidelberg, Germany, 1991; pp. 283–293. [Google Scholar]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001. [Google Scholar]





| Gene Name | Function |
|---|---|
| Phvul.010G024900 | MPPBETA Insulinase (Peptidase family M16) |
| Phvul.010G025000 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G025100 | NB-ARC domain-containing disease resistance |
| Phvul.010G025200 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G025300 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G025400 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G025500 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G025600 | Unknown |
| Phvul.010G025700 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G025800 | Protein of unknown function (DUF506) |
| Phvul.010G025900 | Mitochondrial transcription termination factor |
| Phvul.010G026000 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G026100 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G026200 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G026300 | target of AVRB operation1 |
| Phvul.010G026400 | Disease resistance protein (TIR-NBS-LRR class) |
| Phvul.010G026500 | Unknown |
| Sample | Replicate | Total Reads | Total Reads Mapped | % Reads Mapped |
|---|---|---|---|---|
| “Sierra” MI | Replicate 1 | 29,470,433 | 26,360,691 | 89.44 |
| “Sierra” MI | Replicate 2 | 15,458,316 | 13,419,654 | 86.81 |
| “Sierra” I | Replicate 1 | 21,564,405 | 18,746,890 | 86.93 |
| “Sierra” I | Replicate 2 | 20,849,023 | 18,915,928 | 90.73 |
| crg MI | Replicate 1 | 10,843,510 | 8,978,807 | 82.80 |
| crg MI | Replicate 2 | 19,146,153 | 16,169,339 | 84.45 |
| crg I | Replicate 1 | 30,918,507 | 27,509,416 | 88.97 |
| crg I | Replicate 2 | 27,518,359 | 24,014,203 | 87.27 |
| Gene Name | crg MI | crg I | “Sierra” I | “Sierra” MI |
|---|---|---|---|---|
| Phvul.010G024900 | 0 | 0 | 0.45 | 1.29 |
| Phvul.010G025000 | 0 | 0 | 0 | 0.01 |
| Phvul.010G025100 | 0 | 0 | 0.01 | 0.01 |
| Phvul.010G025200 | 0 | 0 | 0.11 | 0.16 |
| Phvul.010G025300 | 0 | 0 | 0.13 | 0.14 |
| Phvul.010G025400 | 0.038922 | 0.05 | 5.28 | 4.91 |
| Phvul.010G025500 | 0.039883 | 0.01 | 1.76 | 2.71 |
| Phvul.010G025600 | 0.039755 | 0 | 0.44 | 0.94 |
| Phvul.010G025700 | 0.487667 | 0 | 0 | 0 |
| Phvul.010G025800 | 0 | 0.03 | 18.7 | 13.3 |
| Phvul.010G025900 | 0.162343 | 0 | 0.62 | 1.62 |
| Phvul.010G026000 | 0 | 0 | 1.9 | 1.79 |
| Phvul.010G026100 | 0 | 0.02 | 3.16 | 3.43 |
| Phvul.010G026200 | 0 | 0 | 3.12 | 2.87 |
| Phvul.010G026300 | 0 | 0.06 | 0.36 | 0.1 |
| Phvul.010G026400 | 0 | 0.02 | 2.88 | 2.92 |
| Phvul.010G026500 | 0 | 0 | 0.08 | 0 |
| Sample Name | Target Name | Reporter | Cycle Threshold | CT Mean | ΔΔCT | 2−ΔΔCT |
|---|---|---|---|---|---|---|
| 3a | ||||||
| “Sierra” MI | Cons 7 | SYBR | 28.87731361 | |||
| “Sierra” MI | Cons 7 | SYBR | 28.26262856 | 28.64316559 | ||
| “Sierra” MI | Cons 7 | SYBR | 28.78955269 | |||
| “Sierra” I | Cons 7 | SYBR | 28.02788353 | |||
| “Sierra“ I | Cons 7 | SYBR | 28.2238121 | 28.2289753 | ||
| “Sierra” I | Cons 7 | SYBR | 28.43523407 | |||
| “Sierra” MI | Phvul010G025000 | SYBR | 34.58884 | 0 | 1 | |
| “Sierra” MI | Phvul010G025000 | SYBR | 34.19945 | 34.89832 | 0 | 1 |
| “Sierra” MI | Phvul010G025000 | SYBR | 35.90666 | 0 | 1 | |
| “Sierra” I | Phvul010G025000 | SYBR | 35.85514 | 2.053371 | 0.24092 | |
| “Sierra” I | Phvul010G025000 | SYBR | 36.9123 | 36.5375 | 2.053371 | 0.24092 |
| “Sierra” I | Phvul010G025000 | SYBR | 36.84506 | 2.053371 | 0.24092 | |
| 3b | ||||||
| “Sierra” MI | Cons 7 | SYBR | 29.88477135 | |||
| “Sierra” MI | Cons 7 | SYBR | 30.33911514 | 30.19886 | ||
| “Sierra” MI | Cons 7 | SYBR | 30.37268639 | |||
| “Sierra” I | Cons 7 | SYBR | 28.9278698 | |||
| “Sierra” I | Cons 7 | SYBR | 29.36959648 | 29.31236 | ||
| “Sierra” I | Cons 7 | SYBR | 29.63962555 | |||
| “Sierra” MI | Phvul010G025800 | SYBR | 27.765 | 0 | 1 | |
| “Sierra” MI | Phvul010G025800 | SYBR | 26.9359 | 27.19686 | 0 | 1 |
| “Sierra” MI | Phvul010G025800 | SYBR | 26.88967 | 0 | 1 | |
| “Sierra” I | Phvul010G025800 | SYBR | 23.62484 | 2.57595 | 5.96262 | |
| “Sierra” I | Phvul010G025800 | SYBR | 23.68967 | 23.73442 | 2.57595 | 5.96262 |
| “Sierra” I | Phvul010G025800 | SYBR | 23.88874 | 2.57595 | 5.96262 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todd, A.R.; Donofrio, N.; Sripathi, V.R.; McClean, P.E.; Lee, R.K.; Pastor-Corrales, M.; Kalavacharla, V. Marker-Assisted Molecular Profiling, Deletion Mutant Analysis, and RNA-Seq Reveal a Disease Resistance Cluster Associated with Uromyces appendiculatus Infection in Common Bean Phaseolus vulgaris L. Int. J. Mol. Sci. 2017, 18, 1109. https://doi.org/10.3390/ijms18061109
Todd AR, Donofrio N, Sripathi VR, McClean PE, Lee RK, Pastor-Corrales M, Kalavacharla V. Marker-Assisted Molecular Profiling, Deletion Mutant Analysis, and RNA-Seq Reveal a Disease Resistance Cluster Associated with Uromyces appendiculatus Infection in Common Bean Phaseolus vulgaris L. International Journal of Molecular Sciences. 2017; 18(6):1109. https://doi.org/10.3390/ijms18061109
Chicago/Turabian StyleTodd, Antonette R., Nicole Donofrio, Venkateswara R. Sripathi, Phillip E. McClean, Rian K. Lee, Marcial Pastor-Corrales, and Venu (Kal) Kalavacharla. 2017. "Marker-Assisted Molecular Profiling, Deletion Mutant Analysis, and RNA-Seq Reveal a Disease Resistance Cluster Associated with Uromyces appendiculatus Infection in Common Bean Phaseolus vulgaris L." International Journal of Molecular Sciences 18, no. 6: 1109. https://doi.org/10.3390/ijms18061109
APA StyleTodd, A. R., Donofrio, N., Sripathi, V. R., McClean, P. E., Lee, R. K., Pastor-Corrales, M., & Kalavacharla, V. (2017). Marker-Assisted Molecular Profiling, Deletion Mutant Analysis, and RNA-Seq Reveal a Disease Resistance Cluster Associated with Uromyces appendiculatus Infection in Common Bean Phaseolus vulgaris L. International Journal of Molecular Sciences, 18(6), 1109. https://doi.org/10.3390/ijms18061109