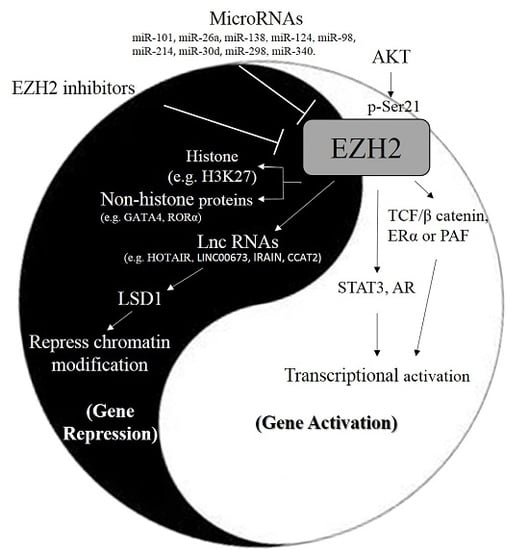
EZH2 in Cancer Progression and Potential Application in Cancer Therapy: A Friend or Foe?
Abstract
:1. Introduction
2. Physiological Functions of EZH2
3. Molecular Regulations of EZH2
3.1. MicroRNAs
3.2. Long Non Coding RNAs
4. Tumor Suppressive Roles of EZH2 in Cancer Progression
5. Oncogenic Roles of EZH2 in Cancer Progression
6. Current Development and Trials of EZH2 Inhibitors
6.1. Pre-Clinical Studies
6.2. Clinical Trials
7. Discussion and Conclusions
Acknowledgments
Conflicts of Interest
References
- Li, L.Y. EZH2: Novel therapeutic target for human cancer. Biomedicine 2014, 4, 1. [Google Scholar] [CrossRef] [PubMed]
- Gall Troselj, K.; Novak Kujundzic, R.; Ugarkovic, D. Polycomb repressive complex’s evolutionary conserved function: The role of EZH2 status and cellular background. Clin. Epigenet. 2016, 8, 55. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The sant domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004, 5, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Carrington, E.A.; Wang, L.; Ketel, C.S.; Miller, E.L.; Jones, R.S.; Simon, J.A. Dominant alleles identify set domain residues required for histone methyltransferase of polycomb repressive complex 2. J. Biol. Chem. 2008, 283, 27757–27766. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, S.J.; Basu, A.; Allis, C.D.; Bernstein, E. Polycomb group proteins: An evolutionary perspective. Trends Genet. 2007, 23, 494–502. [Google Scholar] [CrossRef] [PubMed]
- Simon, J.A.; Kingston, R.E. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Su, I.H.; Dobenecker, M.W.; Dickinson, E.; Oser, M.; Basavaraj, A.; Marqueron, R.; Viale, A.; Reinberg, D.; Wulfing, C.; Tarakhovsky, A. Polycomb group protein EZH2 controls actin polymerization and cell signaling. Cell 2005, 121, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Li, G.; Sarma, K.; Blais, A.; Zavadil, J.; Woodcock, C.L.; Dynlacht, B.D.; Reinberg, D. Ezh1 and EZH2 maintain repressive chromatin through different mechanisms. Mol. Cell 2008, 32, 503–518. [Google Scholar] [CrossRef] [PubMed]
- Bachmann, I.M.; Halvorsen, O.J.; Collett, K.; Stefansson, I.M.; Straume, O.; Haukaas, S.A.; Salvesen, H.B.; Otte, A.P.; Akslen, L.A. EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J. Clin. Oncol. 2006, 24, 268–273. [Google Scholar] [CrossRef] [PubMed]
- Schuettengruber, B.; Cavalli, G. Recruitment of polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009, 136, 3531–3542. [Google Scholar] [CrossRef] [PubMed]
- Aoto, T.; Saitoh, N.; Sakamoto, Y.; Watanabe, S.; Nakao, M. Polycomb group protein-associated chromatin is reproduced in post-mitotic G1 phase and is required for s phase progression. J. Biol. Chem. 2008, 283, 18905–18915. [Google Scholar] [CrossRef] [PubMed]
- Bracken, A.P.; Kleine-Kohlbrecher, D.; Dietrich, N.; Pasini, D.; Gargiulo, G.; Beekman, C.; Theilgaard-Monch, K.; Minucci, S.; Porse, B.T.; Marine, J.C.; et al. The polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007, 21, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Collinson, A.; Collier, A.J.; Morgan, N.P.; Sienerth, A.R.; Chandra, T.; Andrews, S.; Rugg-Gunn, P.J. Deletion of the polycomb-group protein EZH2 leads to compromised self-renewal and differentiation defects in human embryonic stem cells. Cell Rep. 2016, 17, 2700–2714. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Jenner, R.G.; Boyer, L.A.; Guenther, M.G.; Levine, S.S.; Kumar, R.M.; Chevalier, B.; Johnstone, S.E.; Cole, M.F.; Isono, K.; et al. Control of developmental regulators by polycomb in human embryonic stem cells. Cell 2006, 125, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Mochizuki-Kashio, M.; Mishima, Y.; Miyagi, S.; Negishi, M.; Saraya, A.; Konuma, T.; Shinga, J.; Koseki, H.; Iwama, A. Dependency on the polycomb gene EZH2 distinguishes fetal from adult hematopoietic stem cells. Blood 2011, 118, 6553–6561. [Google Scholar] [CrossRef] [PubMed]
- Herviou, L.; Cavalli, G.; Cartron, G.; Klein, B.; Moreaux, J. EZH2 in normal hematopoiesis and hematological malignancies. Oncotarget 2016, 7, 2284–2296. [Google Scholar] [PubMed]
- Yang, X.P.; Jiang, K.; Hirahara, K.; Vahedi, G.; Afzali, B.; Sciume, G.; Bonelli, M.; Sun, H.W.; Jankovic, D.; Kanno, Y.; et al. EZH2 is crucial for both differentiation of regulatory T cells and t effector cell expansion. Sci. Rep. 2015, 5, 10643. [Google Scholar] [CrossRef] [PubMed]
- Su, I.H.; Basavaraj, A.; Krutchinsky, A.N.; Hobert, O.; Ullrich, A.; Chait, B.T.; Tarakhovsky, A. EZH2 controls b cell development through histone H3 methylation and IGH rearrangement. Nat. Immunol. 2003, 4, 124–131. [Google Scholar] [CrossRef] [PubMed]
- Etchegaray, J.P.; Yang, X.; DeBruyne, J.P.; Peters, A.H.; Weaver, D.R.; Jenuwein, T.; Reppert, S.M. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 2006, 281, 21209–21215. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Shen, X.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Wang, G.; Marquez, V.E.; Orkin, S.H.; Pu, W.T. Prc2 directly methylates GATA4 and represses its transcriptional activity. Genes Dev. 2012, 26, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Lee, J.S.; Kim, H.; Kim, K.; Park, H.; Kim, J.Y.; Lee, S.H.; Kim, I.S.; Kim, J.; Lee, M.; et al. Ezh2 generates a methyl degron that is recognized by the DCAF1/DDB1/CUL4 E3 ubiquitin ligase complex. Mol. Cell 2012, 48, 572–586. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Liang, J.; Yang, X.; Wang, Y.; Zhao, Y.; Wu, H.; Sun, L.; Zhang, Y.; Chen, Y.; Li, R.; et al. Integration of estrogen and wnt signaling circuits by the polycomb group protein EZH2 in breast cancer cells. Mol. Cell Biol. 2007, 27, 5105–5119. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.Y.; Jun, S.; Lee, M.; Kim, H.C.; Wang, X.; Ji, H.; McCrea, P.D.; Park, J.I. Paf and EZH2 induce wnt/β-catenin signaling hyperactivation. Mol. Cell 2013, 52, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. Ezh2 oncogenic activity in castration-resistant prostate cancer cells is polycomb-independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.; Kim, M.; Woo, D.H.; Shin, Y.; Shin, J.; Chang, N.; Oh, Y.T.; Kim, H.; Rheey, J.; Nakano, I.; et al. Phosphorylation of EZH2 activates STAT3 signaling via STAT3 methylation and promotes tumorigenicity of glioblastoma stem-like cells. Cancer Cell 2013, 23, 839–852. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.T.; Li, Z.; Wu, Z.; Aau, M.; Guan, P.; Karuturi, R.K.; Liou, Y.C.; Yu, Q. Context-specific regulation of NF-κB target gene expression by EZH2 in breast cancers. Mol. Cell 2011, 43, 798–810. [Google Scholar] [CrossRef] [PubMed]
- Wahid, F.; Shehzad, A.; Khan, T.; Kim, Y.Y. Micrornas: Synthesis, mechanism, function, and recent clinical trials. Biochim. Biophys. Acta 2010, 1803, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Benetatos, L.; Voulgaris, E.; Vartholomatos, G.; Hatzimichael, E. Non-coding rnas and ezh2 interactions in cancer: Long and short tales from the transcriptome. Int. J. Cancer 2013, 133, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, Y.; Ren, K.; Zhao, G.; Yan, K.; Ma, B. Microrna-101 inhibits the metastasis of osteosarcoma cells by downregulation of EZH2 expression. Oncol. Rep. 2014, 32, 2143–2149. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.J.; Ruan, H.J.; He, X.J.; Ma, Y.Y.; Jiang, X.T.; Xia, Y.J.; Ye, Z.Y.; Tao, H.Q. MicroRNA-101 is down-regulated in gastric cancer and involved in cell migration and invasion. Eur. J. Cancer 2010, 46, 2295–2303. [Google Scholar] [CrossRef] [PubMed]
- Varambally, S.; Cao, Q.; Mani, R.S.; Shankar, S.; Wang, X.; Ateeq, B.; Laxman, B.; Cao, X.; Jing, X.; Ramnarayanan, K.; et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science 2008, 322, 1695–1699. [Google Scholar] [CrossRef] [PubMed]
- Smits, M.; Nilsson, J.; Mir, S.E.; van der Stoop, P.M.; Hulleman, E.; Niers, J.M.; de Witt Hamer, P.C.; Marquez, V.E.; Cloos, J.; Krichevsky, A.M.; et al. miR-101 is down-regulated in glioblastoma resulting in EZH2-induced proliferation, migration, and angiogenesis. Oncotarget 2010, 1, 710–720. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Ma, A.; Yang, L.; Hu, H.; Zhu, B.; Shang, D.; Chen, T.; Luo, Y. MicroRNA-26a regulates tumorigenic properties of EZH2 in human lung carcinoma cells. Cancer Genet. 2012, 205, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.C.; Shi, Z.B.; Zhang, Y.T.; Ji, L.; Wang, K.Z.; Duan, D.P.; Dang, X.Q. Downregulation of microRNA-26a is associated with metastatic potential and the poor prognosis of osteosarcoma patients. Oncol. Rep. 2014, 31, 1263–1270. [Google Scholar] [PubMed]
- Zhang, H.; Zhang, H.; Zhao, M.; Lv, Z.; Zhang, X.; Qin, X.; Wang, H.; Wang, S.; Su, J.; Lv, X.; et al. Mir-138 inhibits tumor growth through repression of EZH2 in non-small cell lung cancer. Cell. Physiol. Biochem. 2013, 31, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Zhang, Y.; Jiang, G.; Liu, Z.; Xiang, W.; Chen, X.; Chen, Z.; Zhao, J. miR-138 induces renal carcinoma cell senescence by targeting EZH2 and is downregulated in human clear cell renal cell carcinoma. Oncol. Res. 2013, 21, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wang, C.; Chen, Z.; Jin, Y.; Wang, Y.; Kolokythas, A.; Dai, Y.; Zhou, X. MicroRNA-138 suppresses epithelial-mesenchymal transition in squamous cell carcinoma cell lines. Biochem. J. 2011, 440, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zang, W.; Liu, P.; Wang, Y.; Du, Y.; Chen, X.; Deng, M.; Sun, W.; Wang, L.; Zhao, G.; et al. MicroRNA-124 inhibits cellular proliferation and invasion by targeting ETS-1 in breast cancer. Tumour Biol. 2014, 35, 10897–10904. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Zhang, Z.; Tan, Z.; He, R.; Zeng, X.; Xie, Y.; Li, S.; Tang, G.; Tang, H.; He, X. MicroRNA-124 inhibits proliferation and induces apoptosis by directly repressing EZH2 in gastric cancer. Mol. Cell. Biochem. 2014, 392, 153–159. [Google Scholar] [CrossRef] [PubMed]
- Zheng, F.; Liao, Y.J.; Cai, M.Y.; Liu, Y.H.; Liu, T.H.; Chen, S.P.; Bian, X.W.; Guan, X.Y.; Lin, M.C.; Zeng, Y.X.; et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 2012, 61, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.J.; Chen, J.T.; Hua, L.; Yao, K.H.; Wang, C.Y. Mir-98 inhibits hepatocellular carcinoma cell proliferation via targeting EZH2 and suppressing wnt/β-catenin signaling pathway. Biomed. Pharmacother. 2017, 85, 472–478. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.D.; Yuan, Y.; Zhuang, C.W.; Li, B.L.; Gong, D.J.; Wang, S.G.; Zeng, Z.Y.; Cheng, H.Z. Microrna-98 and microrna-214 post-transcriptionally regulate enhancer of zeste homolog 2 and inhibit migration and invasion in human esophageal squamous cell carcinoma. Mol. Cancer 2012, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Xie, R.; Wu, S.N.; Gao, C.C.; Yang, X.Z.; Wang, H.G.; Zhang, J.L.; Yan, W.; Ma, T.H. Microrna-30d inhibits the migration and invasion of human esophageal squamous cell carcinoma cells via the posttranscriptional regulation of enhancer of zeste homolog 2. Oncol. Rep. 2017, 37, 1682–1690. [Google Scholar] [PubMed]
- Zhou, F.; Chen, J.; Wang, H. MicroRNA-298 inhibits malignant phenotypes of epithelial ovarian cancer by regulating the expression of EZH2. Oncol. Lett. 2016, 12, 3926–3932. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zhang, G.; Lu, B.; Li, J.; Wu, Z.; Ma, H.; Wang, H.; Lian, R. miR-340 impedes the progression of laryngeal squamous cell carcinoma by targeting EZH2. Gene 2016, 577, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ahmad, A.; Kong, D.; Ali, S.; Azmi, A.S.; Li, Y.; Banerjee, S.; Padhye, S.; Sarkar, F.H. Hypoxia induced aggressiveness of prostate cancer cells is linked with deregulated expression of VEGF, IL-6 and mirnas that are attenuated by CDF. PLoS ONE 2012, 7, e43726. [Google Scholar] [CrossRef] [PubMed]
- Bao, B.; Ali, S.; Ahmad, A.; Azmi, A.S.; Li, Y.; Banerjee, S.; Kong, D.; Sethi, S.; Aboukameel, A.; Padhye, S.B.; et al. Hypoxia-induced aggressiveness of pancreatic cancer cells is due to increased expression of VEGF, IL-6 and miR-21, which can be attenuated by CDF treatment. PLoS ONE 2012, 7, e50165. [Google Scholar] [CrossRef] [PubMed]
- Mercer, T.R.; Dinger, M.E.; Mattick, J.S. Long non-coding rnas: Insights into functions. Nat. Rev. Genet. 2009, 10, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Hajjari, M.; Salavaty, A. Hotair: An oncogenic long non-coding rna in different cancers. Cancer Biol. Med. 2015, 12, 1–9. [Google Scholar] [PubMed]
- Gupta, R.A.; Shah, N.; Wang, K.C.; Kim, J.; Horlings, H.M.; Wong, D.J.; Tsai, M.C.; Hung, T.; Argani, P.; Rinn, J.L.; et al. Long non-coding RNA hotair reprograms chromatin state to promote cancer metastasis. Nature 2010, 464, 1071–1076. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.Y.; Wang, J.H.; Wang, J.L.; Ma, C.X.; Wang, X.C.; Liu, F.S. Malat1 as an evolutionarily conserved lncrna, plays a positive role in regulating proliferation and maintaining undifferentiated status of early-stage hematopoietic cells. BMC Genom. 2015, 16, 676. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through EZH2 and interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, Y.; Li, S.; Chen, X.; Jiang, G.; Shen, Z.; Qiao, Y.; Wang, L.; Zheng, P.; Zhang, Y. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting β-catenin via EZH2. Oncotarget 2016, 7, 25668–25682. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Ooi, H.S.; Wu, J.; Chen, J.; Zhang, X.; Tan, S.; Yu, Q.; Li, Y.Y.; Kang, Y.; Li, H.; et al. Malat1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget 2016, 7, 12693–12703. [Google Scholar] [PubMed]
- Wang, X.; Sehgal, L.; Jain, N.; Khashab, T.; Mathur, R.; Samaniego, F. lncRNA malat1 promotes development of mantle cell lymphoma by associating with EZH2. J. Transl. Med. 2016, 14, 346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Z.; Zhao, G.; Zhuang, C.; Shen, Y.Y.; Zhao, W.Y.; Xu, J.; Wang, M.; Wang, C.J.; Tu, L.; Cao, H.; et al. Long non-coding RNA linc00628 functions as a gastric cancer suppressor via long-range modulating the expression of cell cycle related genes. Sci. Rep. 2016, 6, 27435. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.Q.; Zheng, X.D.; Cao, Y.; He, X.D.; Nian, W.Q.; Zeng, X.H.; Liu, X.Y. Long non-coding RNA linc00628 suppresses the growth and metastasis and promotes cell apoptosis in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 275–283. [Google Scholar] [PubMed]
- Huang, M.; Hou, J.; Wang, Y.; Xie, M.; Wei, C.; Nie, F.; Wang, Z.; Sun, M. Long noncoding rna linc00673 is activated by SP1 and exerts oncogenic properties by interacting with LSD1 and EZH2 in gastric cancer. Mol. Ther. 2017, 25, 1014–1026. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Nie, F.; Wang, Y.; Zhang, Z.; Hou, J.; He, D.; Xie, M.; Xu, L.; De, W.; Wang, Z.; et al. LncRNA hoxa11-as promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016, 76, 6299–6310. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, Z.; Fan, R.; Jiang, B.; Chen, X.; Chen, Q.; Nie, F.; Lu, K.; Sun, M. Over-expressed long noncoding RNA HOXA11-as promotes cell cycle progression and metastasis in gastric cancer. Mol. Cancer 2017, 16, 82. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.C.; Li, S.J.; Li, G.; Hua, R.X.; Zhou, X.H.; Li, D.J. Long intergenic noncoding RNA 00511 acts as an oncogene in non-small-cell lung cancer by binding to EZH2 and suppressing p57. Mol. Ther. Nucleic Acids 2016, 5, e385. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.M.; Huang, M.D.; Sun, D.P.; Kong, R.; Xu, T.P.; Xia, R.; Zhang, E.B.; Shu, Y.Q. Long intergenic non-coding RNA 00152 promotes tumor cell cycle progression by binding to EZH2 and repressing P15 and P21 in gastric cancer. Oncotarget 2016, 7, 9773–9787. [Google Scholar] [PubMed]
- Wang, Y.J.; Liu, J.Z.; Lv, P.; Dang, Y.; Gao, J.Y.; Wang, Y. Long non-coding RNA CCAT2 promotes gastric cancer proliferation and invasion by regulating the e-cadherin and LATS2. Am. J. Cancer Res. 2016, 6, 2651–2660. [Google Scholar] [PubMed]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Li, Z.; Wang, W.; Zeng, Y.; Liu, Z.; Qiu, J. Long non-coding RNA h19 increases bladder cancer metastasis by associating with EZH2 and inhibiting e-cadherin expression. Cancer Lett. 2013, 333, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.Y.; Yang, F.; Zhang, Y.L.; Liu, B.; Wang, M.; Hong, X.; Yu, Y.; Zhou, Y.H.; Zeng, H. LncRNA-ancr down-regulation suppresses invasion and migration of colorectal cancer cells by regulating EZH2 expression. Cancer Biomark. 2017, 18, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ancr attenuated the invasion and metastasis of breast cancer. Cell. Death Differ. 2017, 24, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA malat1 regulates alternative splicing by modulating sr splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, R.; Mayeda, A.; Yoshida, M.; Nakagawa, S. Malat1 long non-coding RNA in cancer. Biochim. Biophys. Acta 2016, 1859, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Lian, Y.; Wang, J.; Feng, J.; Ding, J.; Ma, Z.; Li, J.; Peng, P.; De, W.; Wang, K. Long non-coding RNA irain suppresses apoptosis and promotes proliferation by binding to LSD1 and EZH2 in pancreatic cancer. Tumour Biol. 2016, 37, 14929–14937. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lin, Y.; Yang, X.; Wu, X.; He, X. Long noncoding RNA H19 regulates EZH2 expression by interacting with miR-630 and promotes cell invasion in nasopharyngeal carcinoma. Biochem. Biophys. Res. Commun. 2016, 473, 913–919. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Tsirigos, A.; Van Vlierberghe, P.; Nedjic, J.; Trimarchi, T.; Flaherty, M.S.; Ferres-Marco, D.; da Ros, V.; Tang, Z.; Siegle, J.; et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012, 18, 298–301. [Google Scholar] [CrossRef] [PubMed]
- Vanharanta, S.; Shu, W.; Brenet, F.; Hakimi, A.A.; Heguy, A.; Viale, A.; Reuter, V.E.; Hsieh, J.J.; Scandura, J.M.; Massague, J. Epigenetic expansion of VHL-HIF signal output drives multiorgan metastasis in renal cancer. Nat. Med. 2013, 19, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Koyama, D.; Wada, T.; Izumi, T.; Hofgaard, P.O.; Bogen, B.; Furukawa, Y. Phosphorylation-mediated EZH2 inactivation promotes drug resistance in multiple myeloma. J. Clin. Investig. 2015, 125, 4375–4390. [Google Scholar] [CrossRef] [PubMed]
- Mallen-St Clair, J.; Soydaner-Azeloglu, R.; Lee, K.E.; Taylor, L.; Livanos, A.; Pylayeva-Gupta, Y.; Miller, G.; Margueron, R.; Reinberg, D.; Bar-Sagi, D. EZH2 couples pancreatic regeneration to neoplastic progression. Genes Dev. 2012, 26, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Hu, H.; Yang, Y.; Zhou, G.; Shang, Z.; Yang, X.; Sun, K.; Zhan, S.; Yu, Z.; Li, P.; et al. Downregulation of enhancer of zeste homolog 2 (EZH2) is essential for the induction of autophagy and apoptosis in colorectal cancer cells. Genes 2016, 7, 83. [Google Scholar] [CrossRef] [PubMed]
- Song-Bing, H.; Hao, Z.; Jian, Z.; Guo-Qiang, Z.; Tuo, H.; Dai-Wei, W.; Wen, G.; Lin, G.; Yi, Z.; Xiao-Feng, X.; et al. Inhibition of EZH2 expression is associated with the proliferation, apoptosis and migration of SW620 colorectal cancer cells in vitro. Exp. Biol. Med. 2015, 240, 546–555. [Google Scholar]
- Zingg, D.; Debbache, J.; Schaefer, S.M.; Tuncer, E.; Frommel, S.C.; Cheng, P.; Arenas-Ramirez, N.; Haeusel, J.; Zhang, Y.; Bonalli, M.; et al. The epigenetic modifier EZH2 controls melanoma growth and metastasis through silencing of distinct tumour suppressors. Nat. Commun. 2015, 6, 6051. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Yu, Y.; Wu, J.; Bai, J.; Zhao, Y.; Li, C.; Sun, W.; Wang, X. Role of EZH2 in oral squamous cell carcinoma carcinogenesis. Gene 2014, 537, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Mahara, S.; Lee, P.L.; Feng, M.; Tergaonkar, V.; Chng, W.J.; Yu, Q. Hifi-alpha activation underlies a functional switch in the paradoxical role of EZH2/PRC2 in breast cancer. Proc. Natl. Acad. Sci. USA 2016, 113, E3735–E3744. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Okabe, H.; Sakamoto, Y.; Hayashi, H.; Hashimoto, D.; Yokoyama, N.; Sakamoto, K.; Kuroki, H.; Mima, K.; Nitta, H.; et al. Enhancer of zeste homolog 2 (EZH2) promotes progression of cholangiocarcinoma cells by regulating cell cycle and apoptosis. Ann. Surg. Oncol. 2013, 20 (Suppl. S3), S667–S675. [Google Scholar] [CrossRef] [PubMed]
- Hubaux, R.; Thu, K.L.; Coe, B.P.; MacAulay, C.; Lam, S.; Lam, W.L. EZH2 promotes E2F-driven sclc tumorigenesis through modulation of apoptosis and cell-cycle regulation. J. Thorac. Oncol. 2013, 8, 1102–1106. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.Q.; Zhang, L.; Gao, B.S.; Wan, Y.G.; Zhang, X.H.; Chen, B.; Wang, Y.T.; Sun, N.; Fu, Y.W. EZH2 promotes tumor progression by increasing vegf expression in clear cell renal cell carcinoma. Clin. Transl. Oncol. 2015, 17, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Wang, J.; Man, W.Y.; Zhang, Q.W.; Xu, W.G. SiRNA silencing EZH2 reverses cisplatin-resistance of human non-small cell lung and gastric cancer cells. Asian Pac. J. Cancer Prev. 2015, 16, 2425–2430. [Google Scholar] [CrossRef] [PubMed]
- Ougolkov, A.V.; Bilim, V.N.; Billadeau, D.D. Regulation of pancreatic tumor cell proliferation and chemoresistance by the histone methyltransferase enhancer of zeste homologue 2. Clin. Cancer Res. 2008, 14, 6790–6796. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.Y.; Wang, H.; Xiang, P.; Liu, Y.W.; Li, H.Z.; Lei, B.X.; Yu, M.; Qi, S.T. Inhibition of EZH2 reverses chemotherapeutic drug tmz chemosensitivity in glioblastoma. Int. J. Clin. Exp. Pathol. 2014, 7, 6662–6670. [Google Scholar] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cbioportal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cbio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Reva, B.; Antipin, Y.; Sander, C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011, 39, e118. [Google Scholar] [CrossRef] [PubMed]
- Imielinski, M.; Berger, A.H.; Hammerman, P.S.; Hernandez, B.; Pugh, T.J.; Hodis, E.; Cho, J.; Suh, J.; Capelletti, M.; Sivachenko, A.; et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012, 150, 1107–1120. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretic, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Durinck, S.; Stawiski, E.W.; Poirier, J.T.; Modrusan, Z.; Shames, D.S.; Bergbower, E.A.; Guan, Y.; Shin, J.; Guillory, J.; et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012, 44, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; Murray, B.A.; et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar]
- Ciriello, G.; Gatza, M.L.; Beck, A.H.; Wilkerson, M.D.; Rhie, S.K.; Pastore, A.; Zhang, H.; McLellan, M.; Yau, C.; Kandoth, C.; et al. Comprehensive molecular portraits of invasive lobular breast cancer. Cell 2015, 163, 506–519. [Google Scholar] [CrossRef] [PubMed]
- Lefebvre, C.; Bachelot, T.; Filleron, T.; Pedrero, M.; Campone, M.; Soria, J.C.; Massard, C.; Levy, C.; Arnedos, M.; Lacroix-Triki, M.; et al. Mutational profile of metastatic breast cancers: A retrospective analysis. PLoS Med. 2016, 13, e1002201. [Google Scholar] [CrossRef] [PubMed]
- Ahn, S.M.; Jang, S.J.; Shim, J.H.; Kim, D.; Hong, S.M.; Sung, C.O.; Baek, D.; Haq, F.; Ansari, A.A.; Lee, S.Y.; et al. Genomic portrait of resectable hepatocellular carcinomas: Implications of RB1 and FGF19 aberrations for patient stratification. Hepatology 2014, 60, 1972–1982. [Google Scholar] [CrossRef] [PubMed]
- Crompton, B.D.; Stewart, C.; Taylor-Weiner, A.; Alexe, G.; Kurek, K.C.; Calicchio, M.L.; Kiezun, A.; Carter, S.L.; Shukla, S.A.; Mehta, S.S.; et al. The genomic landscape of pediatric ewing sarcoma. Cancer Discov. 2014, 4, 1326–1341. [Google Scholar] [CrossRef] [PubMed]
- Tirode, F.; Surdez, D.; Ma, X.; Parker, M.; Le Deley, M.C.; Bahrami, A.; Zhang, Z.; Lapouble, E.; Grossetete-Lalami, S.; Rusch, M.; et al. Genomic landscape of ewing sarcoma defines an aggressive subtype with co-association of STAG2 and TP53 mutations. Cancer Discov. 2014, 4, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 2013, 499, 43–49. [Google Scholar]
- Ceccarelli, M.; Barthel, F.P.; Malta, T.M.; Sabedot, T.S.; Salama, S.R.; Murray, B.A.; Morozova, O.; Newton, Y.; Radenbaugh, A.; Pagnotta, S.M.; et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 2016, 164, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Johnson, B.E.; Mazor, T.; Hong, C.; Barnes, M.; Aihara, K.; McLean, C.Y.; Fouse, S.D.; Yamamoto, S.; Ueda, H.; Tatsuno, K.; et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 2014, 343, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Morrissy, A.S.; Garzia, L.; Shih, D.J.; Zuyderduyn, S.; Huang, X.; Skowron, P.; Remke, M.; Cavalli, F.M.; Ramaswamy, V.; Lindsay, P.E.; et al. Divergent clonal selection dominates medulloblastoma at recurrence. Nature 2016, 529, 351–357. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas, N. Comprehensive molecular characterization of human colon and rectal cancer. Nature 2012, 487, 330–337. [Google Scholar]
- Giannakis, M.; Mu, X.J.; Shukla, S.A.; Qian, Z.R.; Cohen, O.; Nishihara, R.; Bahl, S.; Cao, Y.; Amin-Mansour, A.; Yamauchi, M.; et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016, 15, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Seshagiri, S.; Stawiski, E.W.; Durinck, S.; Modrusan, Z.; Storm, E.E.; Conboy, C.B.; Chaudhuri, S.; Guan, Y.; Janakiraman, V.; Jaiswal, B.S.; et al. Recurrent R-spondin fusions in colon cancer. Nature 2012, 488, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Van Allen, E.M.; Mouw, K.W.; Kim, P.; Iyer, G.; Wagle, N.; Al-Ahmadie, H.; Zhu, C.; Ostrovnaya, I.; Kryukov, G.V.; O'Connor, K.W.; et al. Somatic ERCC2 mutations correlate with cisplatin sensitivity in muscle-invasive urothelial carcinoma. Cancer Discov. 2014, 4, 1140–1153. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar]
- Kim, P.H.; Cha, E.K.; Sfakianos, J.P.; Iyer, G.; Zabor, E.C.; Scott, S.N.; Ostrovnaya, I.; Ramirez, R.; Sun, A.; Shah, R.; et al. Genomic predictors of survival in patients with high-grade urothelial carcinoma of the bladder. Eur. Urol. 2015, 67, 198–201. [Google Scholar] [CrossRef] [PubMed]
- Al-Ahmadie, H.A.; Iyer, G.; Lee, B.H.; Scott, S.N.; Mehra, R.; Bagrodia, A.; Jordan, E.J.; Gao, S.P.; Ramirez, R.; Cha, E.K.; et al. Frequent somatic CDH1 loss-of-function mutations in plasmacytoid variant bladder cancer. Nat. Genet. 2016, 48, 356–358. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, L.; Ou, Y.; Gao, Z.; Li, E.; Li, X.; Zhang, W.; Wang, J.; Xu, L.; Zhou, Y.; et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature 2014, 509, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Dulak, A.M.; Stojanov, P.; Peng, S.; Lawrence, M.S.; Fox, C.; Stewart, C.; Bandla, S.; Imamura, Y.; Schumacher, S.E.; Shefler, E.; et al. Exome and whole-genome sequencing of esophageal adenocarcinoma identifies recurrent driver events and mutational complexity. Nat. Genet. 2013, 45, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Analysis Working Group: Asan University; BC Cancer Agency; Brigham and Women’s Hospital; Broad Institute; Brown University; Case Western Reserve University; Dana-Farber Cancer Institute; Duke University; Greater Poland Cancer Center; et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017, 541, 169–175. [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef] [PubMed]
- Krauthammer, M.; Kong, Y.; Ha, B.H.; Evans, P.; Bacchiocchi, A.; McCusker, J.P.; Cheng, E.; Davis, M.J.; Goh, G.; Choi, M.; et al. Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma. Nat. Genet. 2012, 44, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Hodis, E.; Watson, I.R.; Kryukov, G.V.; Arold, S.T.; Imielinski, M.; Theurillat, J.P.; Nickerson, E.; Auclair, D.; Li, L.; Place, C.; et al. A landscape of driver mutations in melanoma. Cell 2012, 150, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Hanna, G.J.; Laga, A.C.; Haddad, R.I.; Lorch, J.H.; Hammerman, P.S. Genomic analysis of metastatic cutaneous squamous cell carcinoma. Clin. Cancer Res. 2015, 21, 1447–1456. [Google Scholar] [CrossRef] [PubMed]
- Shain, A.H.; Garrido, M.; Botton, T.; Talevich, E.; Yeh, I.; Sanborn, J.Z.; Chung, J.; Wang, N.J.; Kakavand, H.; Mann, G.J.; et al. Exome sequencing of desmoplastic melanoma identifies recurrent nfkbie promoter mutations and diverse activating mutations in the mapk pathway. Nat. Genet. 2015, 47, 1194–1199. [Google Scholar] [CrossRef] [PubMed]
- Schulze, K.; Imbeaud, S.; Letouze, E.; Alexandrov, L.B.; Calderaro, J.; Rebouissou, S.; Couchy, G.; Meiller, C.; Shinde, J.; Soysouvanh, F.; et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat. Genet. 2015, 47, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Gingras, M.C.; Covington, K.R.; Chang, D.K.; Donehower, L.A.; Gill, A.J.; Ittmann, M.M.; Creighton, C.J.; Johns, A.L.; Shinbrot, E.; Dewal, N.; et al. Ampullary cancers harbor ELF3 tumor suppressor gene mutations and exhibit frequent wnt dysregulation. Cell. Rep. 2016, 14, 907–919. [Google Scholar] [CrossRef] [PubMed]
- Reinhold, W.C.; Sunshine, M.; Liu, H.; Varma, S.; Kohn, K.W.; Morris, J.; Doroshow, J.; Pommier, Y. Cellminer: A web-based suite of genomic and pharmacologic tools to explore transcript and drug patterns in the nci-60 cell line set. Cancer Res. 2012, 72, 3499–3511. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Ley, T.J.; Miller, C.; Ding, L.; Raphael, B.J.; Mungall, A.J.; Robertson, A.; Hoadley, K.; Triche, T.J., Jr.; Laird, P.W.; et al. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013, 368, 2059–2074. [Google Scholar]
- Holmfeldt, L.; Wei, L.; Diaz-Flores, E.; Walsh, M.; Zhang, J.; Ding, L.; Payne-Turner, D.; Churchman, M.; Andersson, A.; Chen, S.C.; et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat. Genet. 2013, 45, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Lohr, J.G.; Stojanov, P.; Lawrence, M.S.; Auclair, D.; Chapuy, B.; Sougnez, C.; Cruz-Gordillo, P.; Knoechel, B.; Asmann, Y.W.; Slager, S.L.; et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc. Natl. Acad. Sci. USA 2012, 109, 3879–3884. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sanada, M.; Shiraishi, Y.; Nowak, D.; Nagata, Y.; Yamamoto, R.; Sato, Y.; Sato-Otsubo, A.; Kon, A.; Nagasaki, M.; et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011, 478, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.; Stransky, N.; McCord, C.L.; Cerami, E.; Lagowski, J.; Kelly, D.; Angiuoli, S.V.; Sausen, M.; Kann, L.; Shukla, M.; et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nat. Commun. 2014, 5, 5006. [Google Scholar] [CrossRef] [PubMed]
- Cancer Genome Atlas Research Network; Kandoth, C.; Schultz, N.; Cherniack, A.D.; Akbani, R.; Liu, Y.; Shen, H.; Robertson, A.G.; Pashtan, I.; Shen, R.; et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013, 497, 67–73. [Google Scholar]
- Crea, F.; Fornaro, L.; Bocci, G.; Sun, L.; Farrar, W.L.; Falcone, A.; Danesi, R. EZH2 inhibition: Targeting the crossroad of tumor invasion and angiogenesis. Cancer Metastasis Rev. 2012, 31, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Neureiter, D.; Wagner, A.; Pichler, M.; Kiesslich, T. The role of polycomb repressive complexes in biliary tract cancer. Expert Opin. Ther. Targets 2015, 19, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Wagner, A.; Stoecklinger, A.; Jakab, M.; Illig, R.; Berr, F.; Pichler, M.; Di Fazio, P.; Ocker, M.; Neureiter, D.; et al. 3-Deazaneplanocin a may directly target putative cancer stem cells in biliary tract cancer. Anticancer Res. 2015, 35, 4697–4705. [Google Scholar] [PubMed]
- Mayr, C.; Wagner, A.; Loeffelberger, M.; Bruckner, D.; Jakab, M.; Berr, F.; Di Fazio, P.; Ocker, M.; Neureiter, D.; Pichler, M.; et al. The BMI1 inhibitor ptc-209 is a potential compound to halt cellular growth in biliary tract cancer cells. Oncotarget 2016, 7, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Amatangelo, M.D.; Garipov, A.; Li, H.; Conejo-Garcia, J.R.; Speicher, D.W.; Zhang, R. Three-dimensional culture sensitizes epithelial ovarian cancer cells to EZH2 methyltransferase inhibition. Cell. Cycle 2013, 12, 2113–2119. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.K.; Tian, X.; LaFrance, L.V.; Duquenne, C.; Suarez, D.P.; Newlander, K.A.; Romeril, S.P.; Burgess, J.L.; Grant, S.W.; Brackley, J.A.; et al. Identification of potent, selective, cell-active inhibitors of the histone lysine methyltransferase EZH2. ACS Med. Chem. Lett. 2012, 3, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Wigle, T.J.; Warholic, N.M.; Sneeringer, C.J.; Allain, C.J.; Klaus, C.R.; Sacks, J.D.; Raimondi, A.; Majer, C.R.; Song, J.; et al. A selective inhibitor of EZH2 blocks H3K27 methylation and kills mutant lymphoma cells. Nat. Chem. Biol. 2012, 8, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.E.; Kuntz, K.W.; Knutson, S.K.; Warholic, N.M.; Keilhack, H.; Wigle, T.J.; Raimondi, A.; Klaus, C.R.; Rioux, N.; Yokoi, A.; et al. Epz011989, a potent, orally-available EZH2 inhibitor with robust in vivo activity. ACS Med. Chem. Lett. 2015, 6, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Qi, W.; Chan, H.; Teng, L.; Li, L.; Chuai, S.; Zhang, R.; Zeng, J.; Li, M.; Fan, H.; Lin, Y.; et al. Selective inhibition of EZH2 by a small molecule inhibitor blocks tumor cells proliferation. Proc. Natl. Acad. Sci. USA 2012, 109, 21360–21365. [Google Scholar] [CrossRef] [PubMed]
- Konze, K.D.; Ma, A.; Li, F.; Barsyte-Lovejoy, D.; Parton, T.; Macnevin, C.J.; Liu, F.; Gao, C.; Huang, X.P.; Kuznetsova, E.; et al. An orally bioavailable chemical probe of the lysine methyltransferases EZH2 and EZH1. ACS Chem. Biol. 2013, 8, 1324–1334. [Google Scholar] [CrossRef] [PubMed]
- Bradley, W.D.; Arora, S.; Busby, J.; Balasubramanian, S.; Gehling, V.S.; Nasveschuk, C.G.; Vaswani, R.G.; Yuan, C.C.; Hatton, C.; Zhao, F.; et al. EZH2 inhibitor efficacy in non-hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem. Biol. 2014, 21, 1463–1475. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Cao, W.; Zhang, J.; Yan, M.; Xu, Q.; Wu, X.; Wan, L.; Zhang, Z.; Zhang, C.; Qin, X.; et al. A covalently bound inhibitor triggers EZH2 degradation through chip-mediated ubiquitination. EMBO J. 2017, 36, 1243–1260. [Google Scholar] [CrossRef] [PubMed]
- Knutson, S.K.; Kawano, S.; Minoshima, Y.; Warholic, N.M.; Huang, K.C.; Xiao, Y.; Kadowaki, T.; Uesugi, M.; Kuznetsov, G.; Kumar, N.; et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-hodgkin lymphoma. Mol. Cancer Ther. 2014, 13, 842–854. [Google Scholar] [CrossRef] [PubMed]
- Vaswani, R.G.; Gehling, V.S.; Dakin, L.A.; Cook, A.S.; Nasveschuk, C.G.; Duplessis, M.; Iyer, P.; Balasubramanian, S.; Zhao, F.; Good, A.C.; et al. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1h-indole-3-carboxamide (cpi-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase i clinical trials for B-cell lymphomas. J. Med. Chem. 2016, 59, 9928–9941. [Google Scholar] [PubMed]
- Zhang, J.; Zheng, Y.G. Sam/sah analogs as versatile tools for sam-dependent methyltransferases. ACS Chem. Biol. 2016, 11, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Harries, M.; Smith, I. The development and clinical use of trastuzumab (herceptin). Endocr. Relat. Cancer 2002, 9, 75–85. [Google Scholar] [CrossRef] [PubMed]
- De Vries, N.A.; Hulsman, D.; Akhtar, W.; de Jong, J.; Miles, D.C.; Blom, M.; van Tellingen, O.; Jonkers, J.; van Lohuizen, M. Prolonged EZH2 depletion in glioblastoma causes a robust switch in cell fate resulting in tumor progression. Cell Rep. 2015, 10, 383–397. [Google Scholar] [CrossRef] [PubMed]

| Target | Subcellular Location | Effects of EZH2 | Roles in Gene Expression | Reference |
|---|---|---|---|---|
| histone H3 | nucleus | tri-methylation of histone H3 at Lys 27 (H3K27me3) | silence | [2] |
| GATA4 | nucleus | methylation of GATA4 to inactivate its functions | silence | [20] |
| RORα | nucleus | methylation-dependent ubiquitination of RORα for its degradation | silence | [21] |
| ERα/ β-catenin | cytoplasm/nucleus | interaction with β-catenin and ERα to link Wnt and estrogen signaling pathways | activation | [22] |
| PAF | nucleus | interaction with PAF to the β-catenin complex to activate Wnt target genes | activation | [23] |
| AKT | cytoplasm/nucleus | phosphorylation of EZH2 at Ser 21 to activate its function | activation | [24] |
| RelA/RelB | cytoplasm/nucleus | interaction with RelA/RelB to activate NF-κB | activation | [26] |
| STAT3 | cytoplasm/nucleus | co-activator of STAT3 | activation | [25] |
| AR | cytoplasm/nucleus | co-activator of AR | activation | [24] |
| MicroRNAs | Effect | Roles in Cancer | Reference |
|---|---|---|---|
| miR-101 | down-regulation of EZH2 expression to inhibit cell proliferation, invasion, and migration abilities in osteosarcoma cells (F5M2) in vitro, gastric cancer cells (MKN-45) in vitro and in vivo (xenograft), prostate cancer cells (SKBr3 and DU145) in vitro and in vivo (xenograft), glioblastoma cells (U87, U118, U251, and U373) and rat GBM cells (C6) in vitro and in vivo (xenograft) | suppressor | [29,30,31,32] |
| miR-26a | inhibition of EZH2 to suppress EMT in human hepatocellular carcinoma; up-regulation of tumor suppressor genes (e.g., DAB2IP and RUNX3) to inhibit cell growth and metastasis in lung carcinoma cells (A549) in vitro and osteosarcoma cells (MG-63 and U20S) in vitro and in tumor tissue samples (in situ) | suppressor | [33,34] |
| miR-138 | inhibition of EZH2 to suppress tumor growth and EMT in NSCLC cells (A549, SPC-A1, SK-MES-1, and H460) and normal human bronchial epithelial cells (16HBE) in vitro and in vivo and in tumor tissue samples (in situ) and squamous cell carcinoma cells (1386Ln and 686Tu) in vitro; targeting EZH2 to induce senescence in human clear cell renal cell carcinoma cells (SN-12) in vitro and in vivo and in tumor tissue samples (in situ) | suppressor | [35,36,37] |
| miR-124 | targeting EZH2 to suppress proliferation in gastric cancer cells (MKN-45, MGC-803, SGC-7901 and AGS) in vitro and in vivo and in tumor tissue samples (in situ); inhibit ROCK2 and EZH2 to repress invasiveness and metastasis in hepatocellular carcinoma cells (Hep3B, Bel-7402, SMMC-7721 and MHCC-LM9) in vitro and in vivo and in tumor tissue samples (in situ) | suppressor | [39,40] |
| miR-98 | inhibition of EZH2 to suppress cells migration and invasion in human esophageal squamous cell carcinoma cells (Eca109) in vitro and in tumor tissue samples (in situ); inhibit cells proliferation via targeting EZH2 to regulate Wnt/β-catenin signaling pathway in hepatocellular carcinoma cells (HepG2) in vitro and in tumor tissue samples (in situ) | suppressor | [41,42] |
| miR-214 | inhibition of EZH2 to suppress migration and invasion in human esophageal squamous cell carcinoma cells (Eca109) in vitro and in tumor tissue samples (in situ) | suppressor | [42] |
| miR-30d | targeting EZH2 to inhibit migration and invasion in human esophageal squamous cell carcinoma cells (ECA109 and KYSE410) in vitro and in tumor tissue samples (in situ) | suppressor | [43] |
| miR-298 | reduction of EZH2 expression to suppress migration and invasion in epithelial ovarian cancer cells (SKOV3 and OVCAR3) in vitro and in tumor tissue samples (in situ) | suppressor | [44] |
| miR-340 | targeting EZH2 to inhibit cancer progression in squamous cell carcinoma cells (Hep-2) in vitro and in tumor tissue samples (in situ) | suppressor | [45] |
| miR-21 | hypoxic state, co-expression with EZH2, IL6, HIF-1α, and VEGF in pancreatic cancer cells (AsPC-1 and MiaPaCa-2) in vitro and in vivo and prostate cancer cells (PC-3 and LNCaP) in vitro | pro-oncogenic | [46,47] |
| miR-210 | hypoxic state, co-expression with EZH2, IL6, HIF-1α, and VEGF in pancreatic cancer cells (AsPC-1 and MiaPaCa-2) in vitro and in vivo and prostate cancer cells (PC-3 and LNCaP) in vitro | pro-oncogenic | [46,47] |
| lncRNAs | Role | Function | Reference |
|---|---|---|---|
| HOTAIR | interaction with EZH2/PRC2 and LSD1 as a repressive chromatin modifier | promoting cancer metastasis via re-localization remodeling of chromatin by PRC2 in many cancer types, including esophageal squamous cell carcinoma cells (KYSE30) in vitro and in tumor tissue samples (in situ), breast cancer cells (MDA-MB-231) in vitro and in vivo (xenograft), and in tumor tissue samples (in situ), and epithelial ovarian cancer cells (SKOV3.ip1 , HO8910-PM, and HEY-A8) in vitro and in vivo (xenograft), and in tumor tissue samples (in situ) | [49,50] |
| MALAT-1 | association with EZH2 | activating EZH2 to suppress p21 and p27 expression and promote cell proliferation in MCL cells (Mino and Jeko-1) in vitro; binding to EZH2 to regulate cancer malignant development in many cancer types, including renal cell carcinoma cells (A-498 and 786-O) in vitro and in tumor tissue samples (in situ), esophageal squamous cell carcinoma cells (TE7) in vitro and in tumor tissue samples (in situ), gastric cancer cells (MKN45 and AGS) in vitro and in tumor tissue samples (in situ), and in MCL cells (Mino and Jeko-1) in vitro and in tumor tissue samples (in situ) | [51,52,53,54,55,68,69] |
| LINC00628 | association with EZH2 | interacting with EZH2 to reduce expression of cell cycle related genes in gastric cancer cells (SGC7901 and MGC-803) in vitro, and tumor size in vivo (xenograft); inhibiting cancer cells growth and metastasis via regulation of Bcl-2/Bax/Caspase-3 signal pathway in breast cancer cells (LCC2 and MCF-7) in vitro and tumor tissue samples (in situ) | [56,57] |
| LINC00673 | a scaffold for interaction with LSD1 and EZH2 | inhibiting expression of KLF2 and LATS2 via association with EZH2 and LSD1 to exert oncogenic functions in gastric cancer cells (BGC823, SGC7901, MGC803, and AGS) in vitro and in vivo (xenograft) | [58] |
| HOXA11-AS | a scaffold for association with PRC2, LSD1, and DNMT1 | promotes cell proliferation, cell cycle progression and metastasis in gastric cancer cells (BGC823 and AGS cells) in vitro, in vivo (xenograft) and in tumor tissue samples (in situ) | [59,60] |
| IRAIN | interaction with EZH2 and LSD1 | interacting with EZH2 and LSD1 to decrease expression of KLF2 and p15 and inhibit apoptosis and cause cycle arrest in pancreatic cancer cells (AsPC-1, BxPC-3, and Panc-1) in vitro and in tumor tissue samples (in situ) | [70] |
| LINC00511 | a scaffold for interaction with EZH2/PRC2 to regulate their localization and functions | suppressing expression of p57 through the association with EZH2 in NSCLC cells (A549 and SPC-A-1) in vitro, in vivo, and in tumor tissue samples (in situ) | [61] |
| LINC00152 | association with EZH2 | promoting gastric cancer cells (SGC-7901 and BGC-823) progression through recruiting EZH2 to suppress p15 and p21 or promote EMT in vitro and metastasis in vivo, and in tumor tissue samples (in situ) | [62] |
| CCAT2 | association with EZH2 and LSD1 | suppressing expression of E-cadherin and LATS2 levels in gastric cancer cells (MKN45 and BGC-823) in vitro and in tumor tissue samples (in situ) | [63] |
| H19 | association with EZH2 | association with EZH2 to activate Wnt/β-catenin and downregulate E-cadherin in bladder cancer cells (RT4 and T24) in vitro and in vivo; interaction with miR-630 to regulate EZH2 level in nasopharyngeal carcinoma cells (CNE2 and HONE1) in vitro and in tumor tissue samples (in situ) | [64,65,71] |
| ANCR | a scaffold for association with EZH2 and CDK1 | decreasing EZH2 to inhibit invasion and metastasis in colorectal cancer cells (SW620) in vitro and in vivo (xenograft); recruiting CDK1 and EZH2 to phosphorylate EZH2 at T345 and T487, hence facilitating EZH2 ubiquitination to degradation, leading to attenuation malignancy in breast cancer cells (MDA-MB-231) in vitro and in vivo (xenograft) and in tumor tissue samples (in situ) | [66,67] |
| Cell type | Model | Function | Roles in Cancer | Reference |
|---|---|---|---|---|
| T-cell acute lymphoblastic leukemia (T-ALL) | CUTLL1, Loucy, Jurkat, MOLT3, HPB-ALL, P12-ichikawa, DND41, CEM2 cell lines (in vitro and in vivo), and tumor tissue samples (in situ) | Loss of EZH2 functions by NOTCH1 pathway and promotes cancer progression. | suppressive | [72] |
| Clear cell renal carcinoma (ccRCC) | 786-O, RFX-631, and OS-RC-2 cell lines (in vitro and in vivo) and in tumor tissue samples (in situ) | Loss of PRC2-mediated histone H3K27me3 activates HIF-driven CXCR4 and increases tumor invasion (suppressive); Overexpression of EZH2 increases VEGF level and cell proliferation (oncogenic). | suppressive or oncogenic | [73,83] |
| Pancreatic cells | EZH2 knockout mice (in vivo) | Loss of EZH2 facilitates K-RasG12D-driven tumor formation | suppressive | [75] |
| Colorectal cancer (CRC) cells | SW620 cell line (in vitro) and in tumor tissue samples (in situ), and RKO and HCT116 cell lines (in vitro) | Inhibition of EZH2 induces autophagy and apoptosis and suppresses cell proliferation and migration. | oncogenic | [76,77] |
| Melanoma | XB2 and Melan-a cell lines (in vitro and in vivo) and in tumor tissue samples (in situ) | EZH2 represses distinct tumor suppressor genes to promote metastasis. | oncogenic | [78] |
| Oral squamous cells carcinoma (OSCC) | Tca8113, Tb, and Ts cell lines (in vitro) and in tumor tissue samples (in situ) | Reducing EZH2 inhibits cell proliferation, migration, metastasis, and induces apoptosis. | oncogenic | [79] |
| Breast cancer | MDA-MB231, HS578T, and BT549 cell lines (in vitro) and in tumor tissue samples (in situ) | PRC2 inhibits expression of MMPs to suppress invasion in normoxia (suppressive). Upon hypoxia, HIF-1α inactivates PRC2 and leads EZH2 to functional switch to EZH2/FoxM1-induced expression of MMPs and invasion (oncogenic). | suppressive or oncogenic | [80] |
| Cholangiocarcinoma cells | RBE and TFK-1 cell lines (in vitro) and in tumor tissue samples (in situ) | Inhibition of EZH2 induces G1 phase arrest, reduces cells growth, and induce apoptosis. | oncogenic | [81] |
| Small cell lung cancer (SCLC) | HTB-175, NCI-H526, HTB-171, HTB-119, and NCI-H524 cell lines (in vitro) | Suppression of EZH2 reduces cells in S or G2/M phases and increases p21 expression. | oncogenic | [82] |
| Cancer Type | Mutation Site | Mutation Type a | Location | Predicted Functional Impact Score (FIS) b | Reference |
|---|---|---|---|---|---|
| Lung adenocarcinoma | A715V | missense | SET domain | 1.19 (low) | [90,91] TCGA data base |
| R34L | missense | non-SET region | 1.50 (low) | ||
| A627E | missense | SET domain | 1.55 (low) | ||
| R502Q | missense | non-SET region | 2.98 (medium) | ||
| E346K | missense | non-SET region | 1.25 (low) | ||
| A622E | missense | non-SET region | 1.55 (low) | ||
| R497Q | missense | non-SET region | 2.98 (medium) | ||
| E341K | missense | non-SET region | 1.25 (low) | ||
| Lung squamous cell carcinoma | E374Q | missense | non-SET region | 2.08 (medium) | [92] TCGA data base |
| S551* | nonsense | non-SET region | - | ||
| Q548E | missense | non-SET region | 1.99 (medium) | ||
| R308L | missense | non-SET region | 2.71 (medium) | ||
| E379Q | missense | non-SET region | 2.08 (medium) | ||
| A345T | missense | non-SET region | 0.55 (neutral) | ||
| S556* | nonsense | non-SET region | - | ||
| Q553E | missense | non-SET region | 1.99 (medium) | ||
| Small cell lung cancer | D185G | missense | non-SET region | 1.04 (low) | [93,94] |
| S40C | missense | non-SET region | - | ||
| Pan-lung cancer | A715V | missense | SET domain | 1.19 (low) | [95] |
| A622E | missense | non-SET region | 1.55 (low) | ||
| R497Q | missense | non-SET region | 2.98 (medium) | ||
| R34L | missense | non-SET region | 1.50 (low) | ||
| E341K | missense | non-SET region | 1.25 (low) | ||
| K510R | missense | non-SET region | 1.87 (low) | ||
| E374Q | missense | non-SET region | 2.08 (medium) | ||
| S551* | nonsense | non-SET region | - | ||
| Q548E | missense | non-SET region | 1.99 (medium) | ||
| G5R | missense | non-SET region | 0.90 (low) | ||
| P262I | missense | non-SET region | - | ||
| R64M | missense | non-SET region | 1.79 (low) | ||
| D186N | missense | non-SET region | 1.50 (low) | ||
| K39E | missense | non-SET region | 1.65 (low) | ||
| R685G | missense | SET domain | - | ||
| H613Q | missense | non-SET region | 1.43 (low) | ||
| K505Yfs*3 | FS del | non-SET region | - | ||
| N310S | missense | non-SET region | 0.41 (neutral) | ||
| R27* | nonsense | non-SET region | - | ||
| Breast invasive carcinoma | S644* | nonsense | SET domain | - | [96,97] TCGA data base |
| E197Rfs*12 | FS del | non-SET region | - | ||
| T718I | missense | SET domain | 0.45 (neutral) | ||
| S639* | nonsense | SET domain | - | ||
| Metastatic breast cancer | A687V | missense | SET domain | 1.14 (low) | [98] |
| L315V | missense | non-SET region | - | ||
| Liver hepatocellular carcinoma | C580* | nonsense | non-SET region | - | [99] TCGA data base |
| E640* | nonsense | SET domain | - | ||
| G395Efs*29 | FS del | non-SET region | - | ||
| I689S | missense | SET domain | −1.22 (neutral) | ||
| Pediatric ewing sarcoma | Y646H | missense | SET domain | 4.61 (high) | [100] |
| Ewing sarcoma | A677G | missense | SET domain | 2.31 (medium) | [101] |
| Y641H | missense | SET domain | 4.61 (high) | ||
| Y641F | missense | SET domain | - | ||
| Clear cell renal cell carcinoma | K6M | missense | non-SET region | −0.46 (neutral) | [102] TCGA data base |
| Q540* | nonsense | non-SET region | - | ||
| D187Gfs*2 | FS ins | non-SET region | - | ||
| Q545* | nonsense | non-SET region | - | ||
| Prostate adenocarcinoma | R16W | missense | non-SET region | 1.04 (low) | TCGA data base |
| Pancreatic adenocarcinoma | R658I | missense | SET domain | 2.44 (medium) | TCGA data base |
| V582A | missense | non-SET region | 1.80 (low) | ||
| A237S | missense | non-SET region | −0.20 (neutral) | ||
| Merged cohort of lower grade glioma (LGG) and glioblastoma multiforme (GBM) | M121I | missense | non-SET region | 2.48 (medium) | [103] |
| Glioblastoma multiforme (GBM) | E396Kfs*22 | FS del | non-SET region | - | [104] TCGA data base |
| K510R | missense | non-SET region | 1.87 (low) | ||
| K515R | missense | non-SET region | 1.87 (low) | ||
| M121I | missense | non-SET region | 2.48 (medium) | ||
| E401Kfs*22 | FS del | non-SET region | - | ||
| Low-grade glioma (LGG) | G11R | missense | non-SET region | 0.69 (neutral) | [105] |
| Medulloblastoma | H706N | missense | SET domain | - | [106] |
| Colorectal adenocarcinoma | C663S | missense | SET domain | 0.53 (neutral) | [107,108,109] TCGA data base |
| R213H | missense | non-SET region | −0.34 (neutral) | ||
| E720K | missense | SET domain | 2.93 (medium) | ||
| E169D | missense | non-SET region | 1.45 (low) | ||
| E725K | missense | SET domain | 2.93 (medium) | ||
| R216Q | missense | non-SET region | 1.04 (low) | ||
| V13A | missense | non-SET region | - | ||
| R16Q | missense | non-SET region | - | ||
| N697D | missense | SET domain | - | ||
| P577L | missense | non-SET region | - | ||
| R354H | missense | non-SET region | - | ||
| L252P | missense | non-SET region | - | ||
| R347W | missense | non-SET region | - | ||
| D202Y | missense | non-SET region | - | ||
| M667T | missense | SET domain | - | ||
| R566C | missense | non-SET region | - | ||
| R313W | missense | non-SET region | - | ||
| S368C | missense | non-SET region | - | ||
| R25Q | missense | non-SET region | |||
| I223F | missense | non-SET region | - | ||
| A255T | missense | non-SET region | - | ||
| N152Ifs*15 | FS del | non-SET region | - | ||
| R347Q | Missense | non-SET region | - | ||
| S368N | missense | non-SET region | - | ||
| D536E | missense | non-SET region | - | ||
| R353C | missense | non-SET region | 2.19 (medium) | ||
| N423T | missense | non-SET region | 0.55 (neutral) | ||
| Bladder urothelial carcinoma | K201E | missense | non-SET region | −0.53 (neutral) | [110,111] |
| S271Y | missense | non-SET region | 2.65 (medium) | ||
| F145Y | missense | non-SET region | - | ||
| Bladder cancer | A596T | missense | non-SET region | 0.64 (neutral) | [112,113] |
| T80Lfs*6 | FS del | non-SET region | - | ||
| A677G | missense | SET domain | 2.31 (medium) | ||
| S639L | missense | SET domain | 0.56 (neutral) | ||
| Esophageal squamous cell carcinoma | V621M | missense | non-SET region | 2.33 (medium) | [114] |
| Esophageal adenocarcinoma | E333Q | missense | non-SET region | 2.19 (medium) | [115] TCGA data base |
| F171S | missense | non-SET region | 2.60 (medium) | ||
| P488S | missense | non-SET region | 1.47 (low) | ||
| D192Y | missense | non-SET region | 0.90 (low) | ||
| Esophagogastric cancer | D192Y | missense | non-SET region | 0.90 (low) | [116] |
| Stomach adenocarcinoma | R18C | missense | non-SET region | 1.94 (medium) | [117] TCGA data base |
| E740K | missense | non-SET region | 1.16 (low) | ||
| M662T | missense | SET domain | 0.33 (neutral) | ||
| S43I | missense | non-SET region | 0.90 (low) | ||
| N668S | missense | SET domain | 0.77 (neutral) | ||
| Cervical squamous cell carcinoma and endocervical adenocarcinoma | P364S | missense | non-SET region | 1.25 (low) | TCGA data base |
| D293H | missense | non-SET region | 2.81 (medium) | ||
| S695L | missense | SET domain | 3.46 (medium) | ||
| Head and neck squamous cell carcinoma | R357W | missense | non-SET region | 1.10 (low) | [118,119] TCGA data base |
| D189N | missense | non-SET region | 0.00 (neutral) | ||
| R362W | missense | non-SET region | 1.10 (low) | ||
| P115S | missense | non-SET region | 2.85 (medium) | ||
| R362Q | missense | non-SET region | −0.29 (neutral) | ||
| R216W | missense | non-SET region | 1.04 (low) | ||
| I264R | missense | non-SET region | 2.62 (medium) | ||
| Y181C | missense | non-SET region | 2.44 (medium) | ||
| S533L | missense | non-SET region | 2.38 (medium) | ||
| Testicular germ cell cancer | K510R | missense | non-SET region | 1.87 (low) | TCGA data base |
| P115T | missense | non-SET region | 2.85 (medium) | ||
| Cholangiocarcinoma | H282N | missense | non-SET region | 2.30 (medium) | TCGA data base |
| Skin cutaneous melanoma | Y641N | missense | SET domain | 4.61 (high) | [120,121] TCGA data base |
| R342Q | missense | non-SET region | 1.15 (low) | ||
| S229L | missense | non-SET region | 1.62 (low) | ||
| Y641F | missense | SET domain | - | ||
| P746S | missense | non-SET region | 0.00 (neutral) | ||
| R34P | missense | non-SET region | 1.50 (low) | ||
| Y641S | missense | SET domain | 4.61 (high) | ||
| S533L | missense | non-SET region | 2.38 (medium) | ||
| R216Q | missense | non-SET region | 1.04 (low) | ||
| P132S | missense | non-SET region | 2.93 (medium) | ||
| R355G | missense | non-SET region | 2.19 (medium) | ||
| P426S | missense | non-SET region | 1.38 (low) | ||
| D142V | missense | non-SET region | 2.90 (medium) | ||
| C530W | missense | non-SET region | 3.57 (high) | ||
| G704S | missense | SET domain | 2.46 (medium) | ||
| A226V | missense | non-SET region | 2.79 (medium) | ||
| T4I | missense | non-SET region | 1.10 (low) | ||
| T4P | missense | non-SET region | 0.41 (neutral) | ||
| Cutaneous squamous cell carcinoma | R685C | missense | SET domain | 4.42 (high) | [122] |
| Y641S | missense | SET domain | 4.61 (high) | ||
| Desmoplastic melanoma | S84L | missense | non-SET region | 0.20 (neutral) | [123] |
| Hepatocellular carcinomas | N670S | missense | SET domain | - | [124] |
| Ampullary carcinoma | Q323K | missense | non-SET region | - | [125] |
| Leukemia | R342Q | missense | non-SET region | 1.15 (low) | [126] |
| Acute myeloid leukemia | E740Afs*24 | FS ins | non-SET region | - | [127] TCGA data base |
| I739Mfs*25 | FS ins | non-SET region | - | ||
| R685H | missense | SET domain | 2.67 (medium) | ||
| E745Afs*24 | FS ins | non-SET region | - | ||
| I744Mfs*25 | FS ins | non-SET region | - | ||
| R690H | missense | SET domain | 2.67 (medium) | ||
| Hypodiploid acute lymphoid leukemia | N670K | missense | SET domain | 1.89 (low) | [128] |
| R679H | missense | SET domain | 2.17 (medium) | ||
| G159R | missense | non-SET region | 2.80 (medium) | ||
| Lymphoid neoplasm diffuse large B-cell lymphoma | Y646F | missense | SET domain | - | TCGA data base |
| Y646S | missense | SET domain | high | ||
| K665R | missense | non-SET region | low | ||
| K665E | missense | non-SET region | low | ||
| D185H | missense | non-SET region | low | ||
| Diffuse large B-Cell lymphoma | Y641F | missense | SET domain | - | [129] |
| Y641N | missense | SET domain | 4.61 (high) | ||
| A687V | missense | SET domain | 1.14 (low) | ||
| Myelodysplasia | K713Efs*12 | FS del | SET domain | - | [130] |
| D659A | missense | SET domain | 2.00 (medium) | ||
| Uterine carcinosarcoma | R608Q | missense | non-SET region | 2.63 (medium) | [131] TCGA data base |
| E59* | nonsense | non-SET region | - | ||
| Uterine corpus endometrial carcinoma | E740K | missense | non-SET region | 1.16 (low) | [132] TCGA data base |
| R497Q | missense | non-SET region | 2.98 (medium) | ||
| Y447* | nonsense | non-SET region | - | ||
| Q540P | missense | non-SET region | 2.25 (medium) | ||
| D233Y | missense | non-SET region | 1.95 (medium) | ||
| E162* | nonsense | non-SET region | - | ||
| F673C | missense | SET domain | 2.69 (medium) | ||
| R78H | missense | non-SET region | 2.14 (medium) | ||
| E246* | nonsense | non-SET region | - | ||
| E396* | nonsense | non-SET region | - | ||
| R349C | missense | non-SET region | 0.90 (low) | ||
| K241Q | missense | non-SET region | 2.51 (medium) | ||
| E721D | missense | SET domain | 4.12 (high) | ||
| R207Q | missense | non-SET region | 0.20 (neutral) |
| Drug | Role | Trial | Stage | Reference |
|---|---|---|---|---|
| 3-Deazaneplanocin A (DZNep) | S-adenosyl-l-homocysteine (SAH) hydrolase inhibitor | many cancer cell lines, such as prostate cancer, brain cancer, and biliary tract cancer cells (EGI-1) | pre-clinical | [133,134,135] |
| GSK926 | SAM-competitive inhibitors of EZH2 | OVCAR10, UPN289 and SKOV3 epithelial ovarian cancer (EOC) cell lines | pre-clinical | [137] |
| GSK343 | SAM-competitive inhibitors of EZH2 | HCC1806, Sk-Br-3 and ZR-75-1 breast cancer cells and LNCaP, PC3 and LNcaP prostate cancer cells | pre-clinical | [138] |
| EPZ-005687 | Inhibitor of EZH2 T641 and A677 mutants | mutant lymphoma cells (heterozygous Tyr 641 or Ala 677) | pre-clinical | [139] |
| EPZ-011989 | selective, oral inhibitor of EZH2 | EZH2 mutant WSU-DLCL2 (Y641F) and DLBCL cell lines in xenograft mouse model | pre-clinical | [140] |
| EI1 | SAM-competitive inhibitors of EZH2 | EZH2 mutat cell lines: WSU-DLCL2 (Y641F), SH-DHL6 (Y641N), and DLBCL cells wild-type EZH2 cell lines: OCI-Y19, GA10, DLBCL, and G401 rhabdoid tumor cells | pre-clinical | [141] |
| UNC-1999 | SAM-competitive inhibitors of EZH2 and EZH1 | MCF7 breast cancer cells, EZH2 mutant DB cells (Y641N) and DLBCL cells, and HEK293T human embryonic kidney cells | pre-clinical | [142] |
| CPI-169 | SAM-competitive inhibitors of EZH2 | lymphoma cell lines, such as GCB, ABC-DLBCL, BL, and MCL cells | pre-clinical | [143] |
| GNA022 | CHIP-mediated ubiquitination and degradation of EZH2 | HN-6 human epithelial cancer cells, A549 lung cancer cells, human head and neck cancer cell lines: UMSCC-12 SCC-25, HN-4, HN-6, HN-12, HN-13, HN-30, Cal-27, KB, and KB/VCR, and breast cancer cell lines: MDA-MB-231 and MDA-MB-468, and SMMC-7721 hepatocyte carcinoma cells | pre-clinical | [144] |
| Tazemetostat (EPZ-6438, E7438) | SAM-competitive inhibitors of EZH2 | 10 clinical studies on going in B-cell non-Hodgkin’s lymphoma, diffuse large B-cell lymphoma, B-cell lymphomas, follicular lymphoma, malignant rhabdoid tumors (MRT), rhabdoid tumors of the kidney (RTK), atypical teratoid rhabdoid tumors (ATRT), synovial sarcoma, epitheliod sarcoma, mesothelioma, advanced solid tumors, selected tumors with rhabdoid features, INI1-negative tumors, malignant rhabdoid tumor of ovary, renal medullary carcinoma | Phase II | - |
| CPI-1205 | SAM-competitive inhibitors of EZH2 | 1 clinical study on going in B-cell lymphomas | Phase I | - |
| GSK2816126 | SAM-competitive inhibitors of EZH2 | 1 clinical study on going in relapsed/refractory diffuse large B cell lymphoma, transformed follicular lymphoma, other non-Hodgkin’s lymphomas, solid tumors and multiple myeloma | Phase I | - |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, K.-S.; Lin, C.-Y.; Liao, T.-W.; Peng, C.-M.; Lee, S.-C.; Liu, Y.-J.; Chan, W.P.; Chou, R.-H. EZH2 in Cancer Progression and Potential Application in Cancer Therapy: A Friend or Foe? Int. J. Mol. Sci. 2017, 18, 1172. https://doi.org/10.3390/ijms18061172
Yan K-S, Lin C-Y, Liao T-W, Peng C-M, Lee S-C, Liu Y-J, Chan WP, Chou R-H. EZH2 in Cancer Progression and Potential Application in Cancer Therapy: A Friend or Foe? International Journal of Molecular Sciences. 2017; 18(6):1172. https://doi.org/10.3390/ijms18061172
Chicago/Turabian StyleYan, Ke-Sin, Chia-Yuan Lin, Tan-Wei Liao, Cheng-Ming Peng, Shou-Chun Lee, Yi-Jui Liu, Wing P. Chan, and Ruey-Hwang Chou. 2017. "EZH2 in Cancer Progression and Potential Application in Cancer Therapy: A Friend or Foe?" International Journal of Molecular Sciences 18, no. 6: 1172. https://doi.org/10.3390/ijms18061172
APA StyleYan, K. -S., Lin, C. -Y., Liao, T. -W., Peng, C. -M., Lee, S. -C., Liu, Y. -J., Chan, W. P., & Chou, R. -H. (2017). EZH2 in Cancer Progression and Potential Application in Cancer Therapy: A Friend or Foe? International Journal of Molecular Sciences, 18(6), 1172. https://doi.org/10.3390/ijms18061172