Diverse Regulation of Vitamin D Receptor Gene Expression by 1,25-Dihydroxyvitamin D and ATRA in Murine and Human Blood Cells at Early Stages of Their Differentiation
Abstract
:1. Introduction
2. Results
2.1. Activation Of Retinoic Acid 4-Hydroxylase (CYP26A1) Gene Expression in Acute Myeloid Leukemia (AML) Cells
2.2. Detection of Transcriptional Variants of Vitamin D Receptor Gene (VDR) and Their Regulation in Response to All-trans-Retinoic Acid (ATRA) in Normal Human Blood Cells
2.3. Detection Of Transcriptional Activities of VDR and Retinoic Acid Receptors (RARs) in Normal Human Blood Cells
2.4. Detection of Transcriptional Variants of VDR in Murine Blood Cells Using 5′-Rapid Amplification of cDNA Ends (RACE) Assay
2.5. Basal Expression of VDR in Murine Kidneys and in Blood Cells
2.6. Regulation of VDR Expression by 1,25D and ATRA in Murine Kidneys and Blood Cells
2.7. Detection of Transcriptional Activities of RARs and VDR in Murine Blood Cells
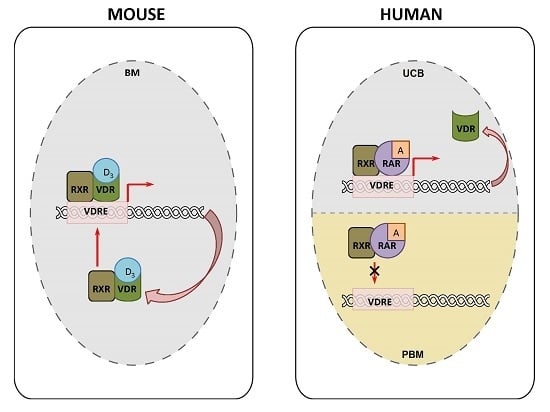
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Lines and Normal Cells
4.3. Sorting of Blood Cells and Flow Cytometry
4.4. 5′-RACE Assay
4.5. cDNA Synthesis and Real-Time PCR
4.6. Statistical Analysis
5. Conlusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| 1,25D | 1,25-Dihydroxyvitamin D |
| AML | Acute myeloid leukemia |
| ATRA | All-trans-retinoic acid |
| BM | Bone marrow |
| BSA | Bovine serum albumin |
| CYP24A1 | 24-Hydroxylase of 1,25D |
| CYP26A1 | Retinoic acid 4-hydroxylase |
| FBS | Fetal bovine serum |
| HSC | Hematopoietic stem cells |
| HSPC | Hematopoeitic stem and progenitor cells |
| GAPDH | Glyceraldehyde 3-phosphate dehydrogenase |
| PCR | Polymerase chain reaction |
| PBM | Peripheral blood mononuclears |
| RA | Retinoic acid |
| RAR | Retinoic acid receptors |
| RARE | Retinoic acid response element |
| RXR | Retinoid X receptors |
| UCB | Umbilical cord blood |
| UTR | Untranslated region |
| VDR | Vitamin D receptor |
| VDRE | Vitamin D response element |
Appendix A



References
- Rhinn, M.; Dollé, P. Retinoic acid signalling during development. Development 2012, 139, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Carlberg, C. The physiology of vitamin D-far more than calcium and bone. Front. Physiol. 2014, 5, 335. [Google Scholar] [CrossRef] [PubMed]
- Chambon, P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996, 10, 940–954. [Google Scholar] [PubMed]
- Kane, M. Analysis, occurrence, and function of 9-cis-retinoic acid. Biochim. Biophys. Acta 2012, 1821, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Baker, A.; McDonnell, D.; Hughes, M.; Crisp, T.; Mangelsdorf, D.; Haussler, M.; Pike, J.; Shine, J.; O’Malley, B. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 3294–3298. [Google Scholar] [CrossRef] [PubMed]
- Aranda, A.; Pascual, A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar] [PubMed]
- Grignani, G.; Ferrucci, P.; Testa, U.; Talamo, G.; Fagioli, M.; Alcalay, M.; Mencarelli, A.; Grignani, F.; Peschle, C.; Nicoletti, I.; et al. The acute promyelocytic leukemia-specific PML-RARα fusion protein inhibits differentiation and promotes survival of myeloid precursor cells. Cell 1993, 74, 423–431. [Google Scholar] [CrossRef]
- Van Etten, E.; Mathieu, C. Immunoregulation by 1,25-dihydroxyvitamin D3: Basic concepts. J. Steroid Biochem. Mol. Biol. 2005, 97, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Brackman, D.; Lund-Johansen, F.; Aarskog, D. Expression of cell surface antigens during the differentiation of HL-60 cells induced by 1,25-dihydroxyvitamin D3, retinoic acid and DMSO. Leuk. Res. 1995, 19, 57–64. [Google Scholar] [CrossRef]
- Tenen, D. Disruption of differentiation in human cancer: AML shows the way. Nat. Rev. Cancer 2003, 3, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Trump, D.; Johnson, C. Vitamin D and acute myeloid leukemia. J. Cancer 2010, 3, 101–107. [Google Scholar] [CrossRef]
- Gocek, E.; Marchwicka, A.; Baurska, H.; Chrobak, A.; Marcinkowska, E. Opposite regulation of vitamin D receptor by ATRA in AML cells susceptible and resistant to vitamin D-induced differentiation. J. Steroid Biochem. Mol. Biol. 2012, 132, 220–226. [Google Scholar] [CrossRef] [PubMed]
- Marchwicka, A.; Cebrat, M.; Łaszkiewicz, A.; Śnieżewski, Ł.; Brown, G.; Marcinkowska, E. Regulation of vitamin D receptor expression by retinoic acid receptor α in acute myeloid leukemia cells. J. Steroid Biochem. Mol. Biol. 2016, 159, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Trump, D.; Johnson, C. Vitamin D in combination cancer treatment. J. Cancer 2010, 1, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Cañete, A.; Cano, E.; Muñoz-Chápuli, R.; Carmona, R. Role of vitamin A/retinoic acid in regulation of embryonic and adult hematopoiesis. Nutrients 2017, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Gritz, E.; Hirschi, K. Specification and function of hemogenic endothelium during embryogenesis. Cell. Mol. Life Sci. 2016, 73, 1547–1567. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Su, C.; Tseng, D.; Ding, F.; Hwang, P. Action of vitamin D and the receptor, VDRa, in calcium handling in zebrafish (Danio rerio). PLoS ONE 2012, 7, e45650. [Google Scholar] [CrossRef] [PubMed]
- Cortes, M.; Chen, M.; Stachura, D.; Liu, S.; Kwan, W.; Wright, F.; Vo, L.; Theodore, L.; Esain, V.; Frost, I.; et al. Developmental vitamin D availability impacts hematopoietic stem cell production. Cell Rep. 2016, 17, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Grande, A.; Montanari, M.; Tagliafico, E.; Manfredini, R.; Marani, T.Z.; Siena, M.; Tenedini, E.; Gallinelli, A.; Ferrari, S. Physiological levels of 1α, 25 dihydroxyvitamin D3 induce the monocytic commitment of CD34+ hematopoietic progenitors. J. Leukoc. Biol. 2002, 71, 641–651. [Google Scholar] [PubMed]
- Crofts, L.; Hancock, M.; Morrison, N.; Eisman, J. Multiple promoters direct the tissue-specific expression of novel N-terminal variant human vitamin D receptor gene transcripts. Proc. Natl. Acad. Sci. USA 1998, 95, 10529–10534. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, K.; Kesterson, R.; Yamamoto, H.; Taketani, Y.; Nishiwaki, E.; Tatsumi, S.; Inoue, Y.; Morita, K.; Takeda, E.; Pike, J. Structural organization of the human vitamin D receptor chromosomal gene and its promoter. Mol. Endocrinol. 1997, 11, 1165–1179. [Google Scholar] [CrossRef] [PubMed]
- Byrne, I.; Flanagan, L.; Tenniswood, M.; Welsh, J. Identification of a hormone-responsive promoter immediately upstream of exon 1c in the human vitamin D receptor gene. Endocrinology 2000, 141, 2829–2836. [Google Scholar] [CrossRef] [PubMed]
- Marchwicka, A.; Corcoran, A.; Berkowska, K.; Marcinkowska, E. Restored expression of vitamin D receptor and sensitivity to 1,25-dihydroxyvitamin D3 in response to disrupted fusion FOP2-FGFR1 gene in acute myeloid leukemia cells. Cell Biosci. 2016, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Wiese, R.; Uhland-Smith, A.; Ross, T.; Prahl, J.; DeLuca, H. Up-regulation of the vitamin D receptor in response to 1,25-dihydroxyvitamin D3 results from ligand-induced stabilization. J. Biol. Chem. 1992, 267, 20082–20086. [Google Scholar] [PubMed]
- Gocek, E.; Kielbinski, M.; Wylob, P.; Kutner, A.; Marcinkowska, E. Side-chain modified vitamin D analogs induce rapid accumulation of VDR in the cell nuclei proportionately to their differentiation-inducing potential. Steroids 2008, 73, 1359–1366. [Google Scholar] [CrossRef] [PubMed]
- Pan, P.; Reddy, K.; Lee, S.; Studzinski, G. Differentiation-related regulation of 1,25-dihydroxyvitamin D3 receptor mRNA in human leukaemia cells HL-60. Cell Prolif. 1991, 24, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Halsall, J.; Osborne, J.; Hutchinson, P.; Pringle, J. In silico analysis of the 5′ region of the vitamin D receptor gene: Functional implications of evolutionary conservation. J. Steroid Biochem. Mol. Biol. 2007, 103, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Gardiner, E.; Esteban, L.; Fong, C.; Allison, S.; Flanagan, J.; Kouzmenko, A.; Eisman, J. Vitamin D receptor b1 and exon 1d: Functional and evolutionary analysis. J. Steroid Biochem. Mol. Biol. 2004, 89–90, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Zella, L.; Kim, S.; Shevde, N.; Pike, J. Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol. Endocrinol. 2006, 20, 1231–1247. [Google Scholar] [CrossRef] [PubMed]
- Zella, L.; Meyer, M.; Nerenz, R.; Lee, S.; Martowicz, M.; Pike, J. Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol. Endocrinol. 2010, 24, 128–147. [Google Scholar] [CrossRef] [PubMed]
- Kahlen, J.; Carlberg, C. Identification of a vitamin D receptor homodimer-type response element in the rat calcitriol 24-hydroxylase gene promoter. Biochem. Biophys. Res. Commun. 1994, 202, 1366–1372. [Google Scholar] [CrossRef] [PubMed]
- Thatcher, J.; Isoherranen, N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin. Drug Metab. Toxicol. 2009, 5, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Ray, W.; Bain, G.; Yao, M.; Gottlieb, D. CYP26, a novel mammalian cytochrome p450, is induced by retinoic acid and defines a new family. J. Biol. Chem. 1997, 272, 18702–18708. [Google Scholar] [CrossRef] [PubMed]
- Loudig, O.; Maclean, G.; Dore, N.; Luu, L.; Petkovich, M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of CYP26A1 inducibility. Biochem. J. 2005, 392, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Ghiaur, G.; Yegnasubramanian, S.; Perkins, B.; Gucwa, J.; Gerber, J.; Jones, R. Regulation of human hematopoietic stem cell self-renewal by the microenvironment’s control of retinoic acid signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 16121–16126. [Google Scholar] [CrossRef] [PubMed]
- Owusu, S.; Ross, A. Retinoid homeostatic gene expression in liver, lung and kidney: Ontogeny and response to vitamin A-retinoic acid (VARA) supplementation from birth to adult age. PLoS ONE 2016, 11, e0145924. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.; Marchwicka, A.; Cunningham, A.; Toellner, K.; Marcinkowska, E. Antagonizing retinoic acid receptors increases myeloid cell production by cultured human hematopoietic stem cells. Arch. Immunol. Ther. Exp. 2017, 65, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Porada, C.; Atala, A.; Almeida-Porada, G. The hematopoietic system in the context of regenerative medicine. Methods 2016, 99, 44–61. [Google Scholar] [CrossRef] [PubMed]
- Gocek, E.; Kielbinski, M.; Baurska, H.; Haus, O.; Kutner, A.; Marcinkowska, E. Different susceptibilities to 1,25-dihydroxyvitamin D3-induced differentiation of aml cells carrying various mutations. Leuk. Res. 2010, 34, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Baurska, H.; Kiełbiński, M.; Biecek, P.; Haus, O.; Jaźwiec, B.; Kutner, A.; Marcinkowska, E. Monocytic differentiation induced by side-chain modified analogs of vitamin D in ex vivo cells from patients with acute myeloid leukemia. Leuk. Res. 2014, 38, 638–647. [Google Scholar] [CrossRef] [PubMed]
- Ablain, J.; de Thé, H. Retinoic acid signaling in cancer: The parable of acute promyelocytic leukemia. Int. J. Cancer 2014, 135, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
- Göttgens, B. Regulatory network control of blood stem cells. Blood 2015, 125, 2614–2620. [Google Scholar] [CrossRef] [PubMed]
- Doulatov, S.; Notta, F.; Laurenti, E.; Dick, J. Hematopoiesis: A human perspective. Cell Stem Cell 2012, 10, 120–136. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, R.; Bischoff-Ferrari, H.; Willett, W. Vitamin D and health: Perspectives from mice and man. J. Bone Miner. Res. 2008, 23, 974–979. [Google Scholar] [CrossRef] [PubMed]
- Christakos, S.; Dhawan, P.; Verstuyf, A.; Verlinden, L.; Carmeliet, G. Vitamin D: Metabolism, molecular mechanism of action, and pleiotropic effects. Physiol. Rev. 2016, 96, 365–408. [Google Scholar] [CrossRef] [PubMed]
- Laszkiewicz, A.; Cebrat, M.; Miazek, A.; Kisielow, P. Complexity of transcriptional regulation within the RAG locus: Identification of a second NWC promoter region within the RAG2 intron. Immunogenetics 2011, 63, 183–187. [Google Scholar] [CrossRef] [PubMed]







© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Janik, S.; Nowak, U.; Łaszkiewicz, A.; Satyr, A.; Majkowski, M.; Marchwicka, A.; Śnieżewski, Ł.; Berkowska, K.; Gabryś, M.; Cebrat, M.; et al. Diverse Regulation of Vitamin D Receptor Gene Expression by 1,25-Dihydroxyvitamin D and ATRA in Murine and Human Blood Cells at Early Stages of Their Differentiation. Int. J. Mol. Sci. 2017, 18, 1323. https://doi.org/10.3390/ijms18061323
Janik S, Nowak U, Łaszkiewicz A, Satyr A, Majkowski M, Marchwicka A, Śnieżewski Ł, Berkowska K, Gabryś M, Cebrat M, et al. Diverse Regulation of Vitamin D Receptor Gene Expression by 1,25-Dihydroxyvitamin D and ATRA in Murine and Human Blood Cells at Early Stages of Their Differentiation. International Journal of Molecular Sciences. 2017; 18(6):1323. https://doi.org/10.3390/ijms18061323
Chicago/Turabian StyleJanik, Sylwia, Urszula Nowak, Agnieszka Łaszkiewicz, Anastasiia Satyr, Michał Majkowski, Aleksandra Marchwicka, Łukasz Śnieżewski, Klaudia Berkowska, Marian Gabryś, Małgorzata Cebrat, and et al. 2017. "Diverse Regulation of Vitamin D Receptor Gene Expression by 1,25-Dihydroxyvitamin D and ATRA in Murine and Human Blood Cells at Early Stages of Their Differentiation" International Journal of Molecular Sciences 18, no. 6: 1323. https://doi.org/10.3390/ijms18061323
APA StyleJanik, S., Nowak, U., Łaszkiewicz, A., Satyr, A., Majkowski, M., Marchwicka, A., Śnieżewski, Ł., Berkowska, K., Gabryś, M., Cebrat, M., & Marcinkowska, E. (2017). Diverse Regulation of Vitamin D Receptor Gene Expression by 1,25-Dihydroxyvitamin D and ATRA in Murine and Human Blood Cells at Early Stages of Their Differentiation. International Journal of Molecular Sciences, 18(6), 1323. https://doi.org/10.3390/ijms18061323