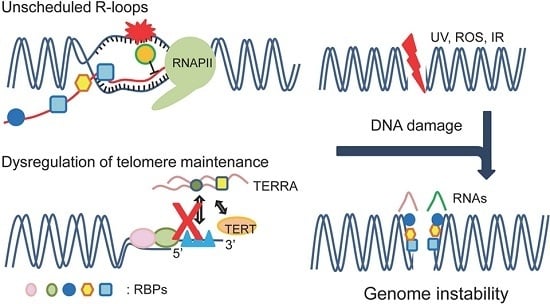
RNA Binding Proteins and Genome Integrity
Abstract
:1. Introduction
2. Role of RBPs in R-Loop Formation
3. Telomere Shortening
4. Roles of Heterogeneous Nuclear Ribonucleoproteins (hnRNP) in Telomere Regulation
5. Roles of RBPs in DNA Damage Responses
6. Possible Roles of Non-Coding RNAs in DNA Damage Responses
7. RNA Modification and DNA Damage Responses
8. Conclusions
Acknowledgments
Conflicts of Interest
References
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Plessmann, U.; Harlander, S.; Daiß, J.-L.; Eichner, N.; Lehmann, G.; Schall, K.; Urlaub, H.; Meister, G. A Compendium of RNA-Binding Proteins that Regulate MicroRNA Biogenesis. Mol. Cell 2017, 66, 270–284. [Google Scholar] [CrossRef] [PubMed]
- Geuens, T.; Bouhy, D.; Timmerman, V. The hnRNP family: Insights into their role in health and disease. Hum. Genet. 2016, 135, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Maziuk, B.; Ballance, H.I.; Wolozin, B. Dysregulation of RNA Binding Protein Aggregation in Neurodegenerative Disorders. Front. Mol. Neurosci. 2017, 10, 89. [Google Scholar] [CrossRef] [PubMed]
- Wurth, L.; Gebauer, F. RNA-binding proteins, multifaceted translational regulators in cancer. BBA Gene Regul. Mech. 2015, 1849, 881–886. [Google Scholar] [CrossRef] [PubMed]
- Hudson, W.H.; Ortlund, E.A. The structure, function and evolution of proteins that bind DNA and RNA. Nat. Rev. Mol. Cell Biol. 2014, 15, 749–760. [Google Scholar] [CrossRef] [PubMed]
- Santos-Pereira, J.M.; Aguilera, A. R loops: New modulators of genome dynamics and function. Nat. Rev. Genet. 2015, 16, 583–597. [Google Scholar] [CrossRef] [PubMed]
- Westover, K.D.; Bushnell, D.A.; Kornberg, R.D. Structural Basis of Transcription: Separation of RNA from DNA by RNA Polymerase II. Science 2004, 303, 1014–1016. [Google Scholar] [CrossRef] [PubMed]
- Drolets, M.; Bi, X.; Lius, L.F. Hypernegative supercoiling of the DNA template during transcription Elongation in Vitro. J. Biol. Chem. 1994, 269, 2068–2074. [Google Scholar]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J.; Gromak, N. Human Senataxin Resolves RNA/DNA Hybrids Formed at Transcriptional Pause Sites to Promote Xrn2-Dependent Termination. Mol. Cell 2011, 42, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.Y.; Clayton, D.A. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem. 1998, 273, 30614–30621. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.; Chedin, F.; Hsieh, C.-L.; Wilson, T.E.; Lieber, M.R. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat. Immunol. 2003, 4, 442–451. [Google Scholar] [CrossRef] [PubMed]
- Chédin, F. Nascent Connections: R-Loops and Chromatin Patterning. Trends Genet. 2016, 32, 828–838. [Google Scholar] [CrossRef] [PubMed]
- Skourti-Stathaki, K.; Proudfoot, N.J. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014, 28, 1384–1396. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. Cotranscriptional processes and their influence on genome stability. Genes Dev. 2006, 1838–1847. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; García-Muse, T. R Loops: From Transcription Byproducts to Threats to Genome Stability. Mol. Cell 2012, 46, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A. The connection between transcription and genomic instability. EMBO J. 2002, 21, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Madireddy, A.; Kosiyatrakul, S.T.; Boisvert, R.A.; Herrera-Moyano, E.; García-Rubio, M.L.; Gerhardt, J.; Vuono, E.; Owen, N.; Yan, Z.; Olson, S.; et al. FANCD2 Facilitates Replication through Common Fragile Sites. Mol. Cell 2016, 64, 388–404. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.A.; Hieter, P.; Stirling, P.C. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 2014, 30, 245–253. [Google Scholar] [CrossRef] [PubMed]
- Marinello, J.; Bertoncini, S.; Aloisi, I.; Cristini, A.; Malagoli Tagliazucchi, G.; Forcato, M.; Sordet, O.; Capranico, G. Dynamic Effects of Topoisomerase I Inhibition on R-Loops and Short Transcripts at Active Promoters. PLoS ONE 2016, 11, e0147053. [Google Scholar] [CrossRef] [PubMed]
- El Hage, A.; French, S.L.; Beyer, A.L.; Tollervey, D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010, 24, 1546–1558. [Google Scholar] [CrossRef] [PubMed]
- Maslon, M.M.; Heras, S.R.; Bellora, N.; Eyras, E.; Cáceres, J.F. The translational landscape of the splicing factor SRSF1 and its role in mitosis. Elife 2014, 2014, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell 2005, 122, 365–378. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Manley, J.L. New talents for an old acquaintance: The SR protein splicing factor ASF/SF2 functions in the maintenance of genome stability. Cell Cycle 2005, 4, 1706–1708. [Google Scholar] [CrossRef] [PubMed]
- Sollier, J.; Stork, C.T.; García-Rubio, M.L.; Paulsen, R.D.; Aguilera, A.; Cimprich, K.A. Transcription-Coupled Nucleotide Excision Repair Factors Promote R-Loop-Induced Genome Instability. Mol. Cell 2014, 56, 777–785. [Google Scholar] [CrossRef] [PubMed]
- Sträßer, K.; Masuda, S.; Mason, P.; Pfannstiel, J.; Oppizzi, M.; Rodriguez-Navarro, S.; Rondón, A.G.; Aguilera, A.; Struhl, K.; Reed, R.; et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 2002, 417, 304–308. [Google Scholar] [CrossRef] [PubMed]
- Rondón, A.G.; Jimeno, S.; García-Rubio, M.; Aguilera, A. Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J. Biol. Chem. 2003, 278, 39037–39043. [Google Scholar] [CrossRef] [PubMed]
- Piruat, J.I.; Aguilera, A.S. A novel yeast gene, THO2, is involved in RNA pol II transcription and provides new evidence for transcriptional elongation-associated recombination. EMBO J. 1998, 17, 4859–4872. [Google Scholar] [CrossRef] [PubMed]
- Aguilera, A.; Klein, H.L. HPR1, a novel yeast gene that prevents intrachromosomal excision recombination, shows carboxy-terminal homology to the Saccharomyces cerevisiae TOP1 gene. Mol. Cell. Biol. 1990, 10, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Huertas, P.; Aguilera, A. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 2003, 12, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.B.; Marı, A.; Moreno, S. The Npl3 hnRNP prevents R-loop- mediated transcription–replication conflicts and genome instability. Genes Dev. 2013, 27, 2445–2458. [Google Scholar]
- Ginno, P.A.; Lim, Y.W.; Lott, P.L.; Korf, I.; Chédin, F. GC skew at the 5′ and 3′ ends of human genes links R-loop formation to epigenetic regulation and transcription termination. Genome Res. 2013, 23, 1590–1600. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, E.J.; Warren, C.L.; Kuehner, J.N.; Panbehi, B.; Ansari, A.Z.; Brow, D.A. Genome-Wide Distribution of Yeast RNA Polymerase II and Its Control by Sen1 Helicase. Mol. Cell 2006, 24, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Mischo, H.E.; Gómez-González, B.; Grzechnik, P.; Rondón, A.G.; Wei, W.; Steinmetz, L.; Aguilera, A.; Proudfoot, N.J. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol. Cell 2011, 41, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Amon, J.D.; Koshland, D. RNase H enables efficient repair of R-loop induced DNA damage. Elife 2016, 5, e20533. [Google Scholar] [CrossRef] [PubMed]
- Ohle, C.; Tesorero, R.; Schermann, G.; Dobrev, N.; Sinning, I.; Fischer, T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001–1013. [Google Scholar] [CrossRef] [PubMed]
- Groh, M.; Gromak, N. Out of Balance: R-loops in Human Disease. PLoS Genet. 2014, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Richard, P.; Manley, J.L. R loops and links to human disease. J. Mol. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- d’Adda di Fagagna, F.; Reaper, P.M.; Clay-Farrace, L.; Fiegler, H.; Carr, P.; von Zglinicki, T.; Saretzki, G.; Carter, N.P.; Jackson, S.P. A DNA damage checkpoint response in telomere-initiated senescence. Nature 2003, 426, 194–198. [Google Scholar] [CrossRef] [PubMed]
- Sfeir, A.; de Lange, T. Removal of shelterin reveals the telomere end-protection problem. Science 2012, 336, 593–597. [Google Scholar] [CrossRef] [PubMed]
- Arnoult, N.; Karlseder, J. Complex interactions between the DNA-damage response and mammalian telomeres. Nat. Struct. Mol. Biol. 2015, 22, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Lazzerini-Denchi, E.; Sfeir, A. Stop pulling my strings–what telomeres taught us about the DNA damage response. Nat. Rev. Mol. Cell Biol. 2016, 17, 364–378. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Telomere-related Genome Instability in Cancer. Cold Spring Harb. Symp. Quant. Biol. 2005, 70, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Maciejowski, J.; de Lange, T. Telomeres in cancer: Tumour suppression and genome instability. Nat. Rev. Mol. Cell Biol. 2017, 18, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-S.; Manche, L.; Xu, R.-M.; Krainer, A.R. hnRNP A1 associates with telomere ends and stimulates telomerase activity. RNA 2006, 12, 1116–1128. [Google Scholar] [CrossRef] [PubMed]
- Flynn, R.L.; Centore, R.C.; O’Sullivan, R.J.; Rai, R.; Tse, A.; Songyang, Z.; Chang, S.; Karlseder, J.; Zou, L. TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 2011, 471, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Takahama, K.; Oyoshi, T. Specific binding of modified RGG domain in tls/fus to G-Quadruplex RNA: Tyrosines in RGG domain recognize 2’-OH of the riboses of loops in G-Quadruplex. J. Am. Chem. Soc. 2013, 135, 18016–18019. [Google Scholar] [CrossRef] [PubMed]
- Takahama, K.; Miyawaki, A.; Shitara, T.; Mitsuya, K.; Morikawa, M.; Hagihara, M.; Kino, K.; Yamamoto, A.; Oyoshi, T. G-Quadruplex DNA- and RNA-Specific-Binding Proteins Engineered from the RGG Domain of TLS/FUS. ACS Chem. Biol. 2015, 10, 2564–2569. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; de Lange, T. Control of telomere length by the human telomeric protein TRF1. Nature 1997, 385, 740–743. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chung, I.K. The splicing factor U2AF65 stabilizes TRF1 protein by inhibiting its ubiquitin-dependent proteolysis. Biochem. Biophys. Res. Commun. 2014, 443, 1124–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Furukawa, K.; Opresko, P.L.; Xu, X.; Bohr, V.A.; Mattson, M.P. TRF2 dysfunction elicits DNA damage responses associated with senescence in proliferating neural cells and differentiation of neurons. J. Neurochem. 2006, 97, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Lopez de Silanes, I.; Stagno d’Alcontres, M.; Blasco, M.A. TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 2010, 1, 33. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.; Collins, K. Purification of Human Telomerase Complexes Identifies Factors Involved in Telomerase Biogenesis and Telomere Length Regulation. Mol. Cell 2007, 28, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Pont, A.R.; Sadri, N.; Hsiao, S.J.; Smith, S.; Schneider, R.J. mRNA Decay Factor AUF1 Maintains Normal Aging, Telomere Maintenance, and Suppression of Senescence by Activation of Telomerase Transcription. Mol. Cell 2012, 47, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Grammatikakis, I.; Zhang, P.; Panda, A.C.; Kim, J.; Maudsley, S.; Abdelmohsen, K.; Yang, X.; Martindale, J.L.; Motiño, O.; Hutchison, E.R.; et al. Alternative Splicing of Neuronal Differentiation Factor TRF2 Regulated by HNRNPH1/H2. Cell Rep. 2016, 15, 926–934. [Google Scholar] [CrossRef] [PubMed]
- Zenklusen, D.; Vinciguerra, P.; Strahm, Y.; Stutz, F. The yeast hnRNP-Like proteins Yra1p and Yra2p participate in mRNA export through interaction with Mex67p. Mol. Cell. Biol. 2001, 21, 4219–4232. [Google Scholar] [CrossRef] [PubMed]
- Gavaldá, S.; Santos-Pereira, J.M.; García-Rubio, M.L.; Luna, R.; Aguilera, A. Excess of Yra1 RNA-Binding Factor Causes Transcription-Dependent Genome Instability, Replication Impairment and Telomere Shortening. PLoS Genet. 2016, 12, e1005966. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, Y.; Mao, P.; Li, F.; Han, X.; Zhang, Y.; Jiang, S.; Chen, Y.; Huang, J.; Liu, D.; et al. Cold-inducible RNA-binding protein CIRP/hnRNP A18 regulates telomerase activity in a temperature-dependent manner. Nucleic Acids Res. 2016, 44, 761–775. [Google Scholar] [CrossRef] [PubMed]
- Ciccia, A.; Elledge, S.J. The DNA Damage Response: Making It Safe to Play with Knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, M.; Lambert, S.; Carreira, A.; Amor-Guéret, M.; Vagner, S. DNA damage: RNA-binding proteins protect from near and far. Trends Biochem. Sci. 2014, 39, 141–149. [Google Scholar] [CrossRef] [PubMed]
- Dutertre, M.; Vagner, S. DNA-Damage Response RNA-Binding Proteins (DDRBPs): Perspectives from a New Class of Proteins and Their RNA Targets. J. Mol. Biol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Montecucco, A.; Biamonti, G. Pre-mRNA processing factors meet the DNA damage response. Front. Genet. 2013, 4, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Adamson, B.; Smogorzewska, A.; Sigoillot, F.D.; King, R.W.; Elledge, S.J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 2012, 14, 318–328. [Google Scholar] [CrossRef] [PubMed]
- Anantha, R.W.; Alcivar, A.L.; Ma, J.; Cai, H.; Simhadri, S.; Ule, J.; König, J.; Xia, B. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS ONE 2013, 8, e61368. [Google Scholar] [CrossRef] [PubMed]
- Rajesh, C.; Baker, D.K.; Pierce, A.J.; Pittman, D.L. The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion. Nucleic Acids Res. 2011, 39, 132–145. [Google Scholar] [CrossRef] [PubMed]
- Ha, K.; Takeda, Y.; Dynan, W.S. Sequences in PSF/SFPQ mediate radioresistance and recruitment of PSF/SFPQ-containing complexes to DNA damage sites in human cells. DNA Repair 2011, 10, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Hong, Z.; Jiang, J.; Ma, J.; Dai, S.; Xu, T.; Li, H.; Yasui, A. The Role of hnRPUL1 Involved in DNA Damage Response Is Related to PARP1. PLoS ONE 2013, 8, e60208. [Google Scholar] [CrossRef] [PubMed]
- Lossaint, G.; Larroque, M.; Ribeyre, C.; Bec, N.; Larroque, C.; Décaillet, C.; Gari, K.; Constantinou, A. FANCD2 Binds MCM Proteins and Controls Replisome Function upon Activation of S Phase Checkpoint Signaling. Mol. Cell 2013, 51, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Mastrocola, A.S.; Kim, S.H.; Trinh, A.T.; Rodenkirch, L.A.; Tibbetts, R.S. The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 2013, 288, 24731–24741. [Google Scholar] [CrossRef] [PubMed]
- Polo, S.E.; Blackford, A.N.; Chapman, J.R.; Baskcomb, L.; Gravel, S.; Rusch, A.; Thomas, A.; Blundred, R.; Smith, P.; Kzhyshkowska, J.; et al. Regulation of DNA-End Resection by hnRNPU-like Proteins Promotes DNA Double-Strand Break Signaling and Repair. Mol. Cell 2012, 45, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Rulten, S.L.; Rotheray, A.; Green, R.L.; Grundy, G.J.; Moore, D.A.Q.; Gomez-Herreros, F.; Hafezparast, M.; Caldecott, K.W. PARP-1 dependent recruitment of the amyotrophic lateral sclerosis-associated protein FUS/TLS to sites of oxidative DNA damage. Nucleic Acids Res. 2014, 42, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Gaudreault, I.; Guay, D.; Lebel, M. YB-1 promotes strand separation in vitro of duplex DNA containing either mispaired bases or cisplatin modifications, exhibits endonucleolytic activities and binds several DNA repair proteins. Nucleic Acids Res. 2004, 32, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-W.; Mai, R.-T.; Fang, W.-H.; Lin, C.-C.; Chiu, C.-C.; Wu Lee, Y.-H. YB-1 disrupts mismatch repair complex formation, interferes with MutSα recruitment on mismatch and inhibits mismatch repair through interacting with PCNA. Oncogene 2014, 33, 5065–5077. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.R.; Selyutina, A.A.; Buldakov, I.A.; Evdokimova, V.; Ovchinnikov, L.P.; Sorokin, A.V. The proteolytic YB-1 fragment interacts with DNA repair machinery and enhances survival during DNA damaging stress. Cell Cycle 2013, 12, 3791–3803. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Kuhne, W.W.; Kulharya, A.; Hudson, F.Z.; Ha, K.; Cao, Z.; Dynan, W.S. Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 2009, 37, 6746–6753. [Google Scholar] [CrossRef] [PubMed]
- Krietsch, J.; Caron, M.-C.; Gagné, J.-P.; Ethier, C.; Vignard, J.; Vincent, M.; Rouleau, M.; Hendzel, M.J.; Poirier, G.G.; Masson, J.-Y. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012, 40, 10287–10301. [Google Scholar] [CrossRef] [PubMed]
- Salton, M.; Lerenthal, Y.; Wang, S.-Y.; Chen, D.J.; Shiloh, Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 2010, 9, 1568–1576. [Google Scholar] [CrossRef] [PubMed]
- Jaafar, L.; Li, Z.; Li, S.; Dynan, W.S. SFPQ·NONO and XLF function separately and together to promote DNA double-strand break repair via canonical nonhomologous end joining. Nucleic Acids Res. 2017, 45, 1848–1859. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, L.; Su, S.C.; Quinn, E.J.; Sasaki, M.; Jimenez, J.C.; Mackenzie, I.R.A.; Huang, E.J.; Tsai, L. Interaction of FUS and HDAC1 regulates DNA damage response and repair in neurons. Nat. Neurosci. 2013, 16, 1383–1391. [Google Scholar] [CrossRef] [PubMed]
- Simon, N.E.; Yuan, M.; Kai, M. RNA-binding protein RBM14 regulates dissociation and association of non-homologous end joining proteins. Cell Cycle 2017. [Google Scholar] [CrossRef] [PubMed]
- Kai, M. Roles of RNA-Binding Proteins in DNA Damage Response. Int. J. Mol. Sci. 2016, 17, 310. [Google Scholar] [CrossRef] [PubMed]
- Shin, K.-H.; Kim, R.H.; Kang, M.K.; Kim, R.H.; Kim, S.G.; Lim, P.K.; Yochim, J.M.; Baluda, M.A.; Park, N.-H. p53 promotes the fidelity of DNA end-joining activity by, in part, enhancing the expression of heterogeneous nuclear ribonucleoprotein G. DNA Repair 2007, 6, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Song, E.J.; Werner, S.L.; Neubauer, J.; Stegmeier, F.; Aspden, J.; Rio, D.; Harper, J.W.; Elledge, S.J.; Kirschner, M.W.; Rape, M. The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 2010, 24, 1434–1447. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.-H.; Kao, D.-I.; Chan, S.-P.; Kao, T.-C.; Lin, J.-Y.; Cheng, S.-C. Functional links between the Prp19-associated complex, U4/U6 biogenesis, and spliceosome recycling. RNA 2006, 12, 765–774. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.-P.; Kao, D.-I.; Tsai, W.-Y.; Cheng, S.-C. The Prp19p-Associated Complex in Spliceosome Activation. Science 2003, 302, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.-P.; Cheng, S.-C. The Prp19-associated Complex Is Required for Specifying Interactions of U5 and U6 with Pre-mRNA during Spliceosome Activation. J. Biol. Chem. 2005, 280, 31190–31199. [Google Scholar] [CrossRef] [PubMed]
- Maréchal, A.; Li, J.-M.; Ji, X.Y.; Wu, C.-S.; Yazinski, S.A.; Nguyen, H.D.; Liu, S.; Jiménez, A.E.; Jin, J.; Zou, L. PRP19 Transforms into a Sensor of RPA-ssDNA after DNA Damage and Drives ATR Activation via a Ubiquitin-Mediated Circuitry. Mol. Cell 2014, 53, 235–246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Kaur, R.; Akhter, S.; Legerski, R.J. Cdc5L interacts with ATR and is required for the S-phase cell-cycle checkpoint. EMBO Rep. 2009, 10, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Clark, M.B.; Amaral, P.P.; Schlesinger, F.J.; Dinger, M.E.; Taft, R.J.; Rinn, J.L.; Ponting, C.P.; Stadler, P.F.; Morris, K.V.; Morillon, A.; et al. The Reality of Pervasive Transcription. PLoS Biol. 2011, 9, e1000625. [Google Scholar] [CrossRef] [PubMed]
- Castel, S.E.; Martienssen, R.A. RNA interference in the nucleus: Roles for small RNAs in transcription, epigenetics and beyond. Nat. Rev. Genet. 2013, 14, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Nadal-Ribelles, M.; Solé, C.; Xu, Z.; Steinmetz, L.M.; de Nadal, E.; Posas, F. Control of Cdc28 CDK1 by a Stress-Induced lncRNA. Mol. Cell 2014, 53, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Huarte, M.; Guttman, M.; Feldser, D.; Garber, M.; Koziol, M.J.; Kenzelmann-Broz, D.; Khalil, A.M.; Zuk, O.; Amit, I.; Rabani, M.; et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 2010, 142, 409–419. [Google Scholar] [CrossRef] [PubMed]
- Kajita, K.; Kuwano, Y.; Satake, Y.; Kano, S.; Kurokawa, K.; Akaike, Y.; Masuda, K.; Nishida, K.; Rokutan, K. Ultraconserved region-containing Transformer 2β4 controls senescence of colon cancer cells. Oncogenesis 2016, 5, e213. [Google Scholar] [CrossRef] [PubMed]
- Meers, C.; Keskin, H.; Storici, F. DNA repair by RNA: Templated, or not templated, that is the question. DNA Repair 2016, 44, 17–21. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.-G.; Qi, Y. RNA-directed repair of DNA double-strand breaks. DNA Repair 2015, 32, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Connerty, P.; Ahadi, A.; Hutvagner, G. RNA Binding Proteins in the miRNA Pathway. Int. J. Mol. Sci. 2016, 17, 31. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Danielsen, J.M.R.; Yang, Y.G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [PubMed]
- Keskin, H.; Shen, Y.; Huang, F.; Patel, M.; Yang, T.; Ashley, K.; Mazin, A.V.; Storici, F. Transcript-RNA-templated DNA recombination and repair. Nature 2014, 515, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Nandi, P.; Taylor, M.B.; Stuckey, S.; Bhadsavle, H.P.; Weiss, B.; Storici, F. RNA-driven genetic changes in bacteria and in human cells. Mutat. Res. 2011, 717, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Laurent, B.; Hsu, C.-H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef] [PubMed]


© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nishida, K.; Kuwano, Y.; Nishikawa, T.; Masuda, K.; Rokutan, K. RNA Binding Proteins and Genome Integrity. Int. J. Mol. Sci. 2017, 18, 1341. https://doi.org/10.3390/ijms18071341
Nishida K, Kuwano Y, Nishikawa T, Masuda K, Rokutan K. RNA Binding Proteins and Genome Integrity. International Journal of Molecular Sciences. 2017; 18(7):1341. https://doi.org/10.3390/ijms18071341
Chicago/Turabian StyleNishida, Kensei, Yuki Kuwano, Tatsuya Nishikawa, Kiyoshi Masuda, and Kazuhito Rokutan. 2017. "RNA Binding Proteins and Genome Integrity" International Journal of Molecular Sciences 18, no. 7: 1341. https://doi.org/10.3390/ijms18071341
APA StyleNishida, K., Kuwano, Y., Nishikawa, T., Masuda, K., & Rokutan, K. (2017). RNA Binding Proteins and Genome Integrity. International Journal of Molecular Sciences, 18(7), 1341. https://doi.org/10.3390/ijms18071341