TRAIL, Wnt, Sonic Hedgehog, TGFβ, and miRNA Signalings Are Potential Targets for Oral Cancer Therapy
Abstract
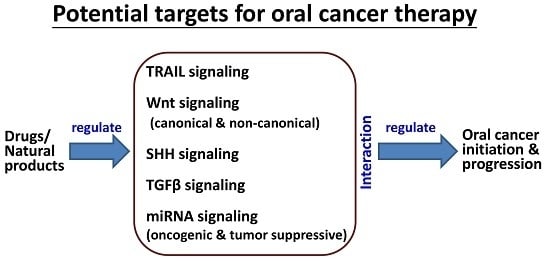
:1. Introduction
2. TRAIL-Induced Intracellular Signaling in Oral Cancer
2.1. TRAIL May Be a Selective Killing Ligand for Cancer Cells
2.2. TRAIL Receptor-Inducible Agents for Targeting Therapies for Oral Cancer
3. Wnt Signaling and Oral Cancer
3.1. Canonical Wnt Pathway in Oral Cancer Cells
3.2. Non-Canonical Wnt Pathway in Oral Cancer Cells
3.3. Wnt Pathway as the Target for Oral Cancer Therapy
4. SHH Signaling and Oral Cancer
5. TGFβ Signaling and Oral Cancer
6. miRNAs and Oral Cancer
6.1. Targets of Oncogenic miRNAs for Oral Cancer Cells
6.1.1. Akt and miR-31
6.1.2. STAT3 and miR-21
6.1.3. PUMA and miR-222
6.2. Targets of Tumor Suppressor miRNAs for Oral Cancer Cells
6.2.1. Neuropilin-1 (NP-1; NRP1) and miR-338
6.2.2. Forkhead Box C1 (FOXC1) and miR-639
6.2.3. Protein Kinase CI (PRKCI) and miR-219
6.2.4. WNT7B, miR-329, and miR-410
6.2.5. Heat Shock Proteins (HSP) and miR-27a
6.2.6. Estrogen-Related Receptor α (ERRα; ESRRA) and miR-125a
6.2.7. Epidermal Growth Factor-Like Domain 7 (EGFL7) and miR-126
6.2.8. Insulin-Like Growth Factor I Receptor (IGF1R) and miR-99a
6.2.9. G-Protein-Coupled Receptor Kinase-Interacting Protein 1 (GIT1) and miR-491-5p
6.2.10. C-X-C Motif Chemokine Receptor 4 (CXCR4) and miR-9
6.2.11. DKK2 and miR-21
7. Interactions between TRAIL, Wnt, SHH, TGFβ, and miRNA Signaling Proteins in Cancer Cells
7.1. TRAIL-Induced Apoptosis and ER Stress
7.2. miRNA and TRAIL Signaling
7.3. Other Complex Crosstalk
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| TRAIL | Tumor necrosis factor-related apoptosis-inducing ligand |
| Wnt | Wingless type MMTV integration site family |
| SHH | Sonic hedgehog |
| TGFβ | Transforming growth factor β |
| DR | Death receptors |
| OSCC | Oral squamous cell carcinomas |
| ERK | Extracellular signal regulated kinase |
| MAPK | Mitogen-activated protein kinase |
| HNSCC | Head and neck squamous cell carcinomas |
| EMT | Epithelial to mesenchymal transition |
| ECM | Extracellular matrix |
| ER stress | Endoplasmic reticulum stress |
References
- Song, K.; Chen, Y.; Goke, R.; Wilmen, A.; Seidel, C.; Goke, A.; Hilliard, B.; Chen, Y. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) is an inhibitor of autoimmune inflammation and cell cycle progression. J. Exp. Med. 2000, 191, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Merino, D.; Lalaoui, N.; Morizot, A.; Solary, E.; Micheau, O. TRAIL in cancer therapy: Present and future challenges. Expert Opin. Ther. Targets 2007, 11, 1299–1314. [Google Scholar] [CrossRef] [PubMed]
- Suzuki-Karasaki, Y.; Suzuki-Karasaki, M.; Uchida, M.; Ochiai, T. Depolarization controls TRAIL-sensitization and tumor-selective killing of cancer cells: Crosstalk with ROS. Front. Oncol. 2014, 4, 128. [Google Scholar] [CrossRef] [PubMed]
- Yurovsky, V.V. Molecular cross-talk between the TRAIL and TGF-β pathways in human lung fibroblasts. Arthritis Res. Ther. 2003, 5, 64. [Google Scholar] [CrossRef]
- Cano-Gonzalez, A.; Lopez-Rivas, A. Opposing roles of TGF-β and EGF in the regulation of TRAIL-induced apoptosis in human breast epithelial cells. Biochim. Biophys. Acta 2016, 1863, 2104–2114. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.; Fu, E. Crosstalk between Shh and TGF-β signaling in cyclosporine-enhanced cell proliferation in human gingival fibroblasts. PLoS ONE 2013, 8, e70128. [Google Scholar] [CrossRef] [PubMed]
- Yoo, Y.A.; Kang, M.H.; Kim, J.S.; Oh, S.C. Sonic hedgehog signaling promotes motility and invasiveness of gastric cancer cells through TGF-β-mediated activation of the ALK5-Smad 3 pathway. Carcinogenesis 2008, 29, 480–490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Tian, X.J.; Xing, J. Signal transduction pathways of EMT induced by TGF-β, SHH, and WNT and their crosstalks. J. Clin. Med. 2016, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Castellone, M.D.; Laukkanen, M.O. TGF-β1, WNT, and SHH signaling in tumor progression and in fibrotic diseases. Front. Biosci. 2017, 9, 31–45. [Google Scholar]
- Johnstone, R.W.; Frew, A.J.; Smyth, M.J. The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat. Rev. Cancer 2008, 8, 782–798. [Google Scholar] [CrossRef] [PubMed]
- Dimberg, L.Y.; Anderson, C.K.; Camidge, R.; Behbakht, K.; Thorburn, A.; Ford, H.L. On the TRAIL to successful cancer therapy? Predicting and counteracting resistance against TRAIL-based therapeutics. Oncogene 2013, 32, 1341–1350. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.S. TRAIL as a target in anti-cancer therapy. Cancer Lett. 2009, 285, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.J.; Shin, J.A.; Lee, S.O.; Kwon, K.H.; Cho, S.D. Extracellular signalregulated kinase inhibition is required for methanol extract of Smilax china L. induced apoptosis through death receptor 5 in human oral mucoepidermoid carcinoma cells. Mol. Med. Rep. 2014, 9, 663–668. [Google Scholar] [PubMed]
- Huong, L.D.; Shin, J.A.; Choi, E.S.; Cho, N.P.; Kim, H.M.; Leem, D.H.; Cho, S.D. β-Phenethyl isothiocyanate induces death receptor 5 to induce apoptosis in human oral cancer cells via p38. Oral Dis. 2012, 18, 513–519. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Huang, Y.; Li, Y.; Pu, L.; Xia, F.; Jiang, C.; Liu, H.; Jiang, Z. Glycosylation inhibitor 2-deoxy-d-glucose sensitizes oral cancer cells to TRAIL-induced apoptosis. Nan Fang Yi Ke Da Xue Xue Bao 2013, 33, 524–527. [Google Scholar] [PubMed]
- Yeh, C.C.; Deng, Y.T.; Sha, D.Y.; Hsiao, M.; Kuo, M.Y. Suberoylanilide hydroxamic acid sensitizes human oral cancer cells to TRAIL-induced apoptosis through increase DR5 expression. Mol. Cancer Ther. 2009, 8, 2718–2725. [Google Scholar] [CrossRef] [PubMed]
- Kok, S.H.; Yeh, C.C.; Chen, M.L.; Kuo, M.Y. Esculetin enhances TRAIL-induced apoptosis through DR5 upregulation in human oral cancer SAS cells. Oral Oncol. 2009, 45, 1067–1072. [Google Scholar] [CrossRef] [PubMed]
- Itashiki, Y.; Harada, K.; Ferdous, T.; Yoshida, H. Effects of tumor necrosis factor-related apoptosis-inducing ligand alone and in combination with fluoropyrimidine anticancer agent, S-1, on tumor growth of human oral squamous cell carcinoma xenografts in nude mice. Anticancer Res. 2007, 27, 2365–2375. [Google Scholar] [PubMed]
- Chen, J.J.; Mikelis, C.M.; Zhang, Y.; Gutkind, J.S.; Zhang, B. TRAIL induces apoptosis in oral squamous carcinoma cells: A crosstalk with oncogenic Ras regulated cell surface expression of death receptor 5. Oncotarget 2013, 4, 206–217. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Habas, R. Wnt signal transduction pathways. Organogenesis 2008, 4, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Reya, T.; Clevers, H. Wnt signalling in stem cells and cancer. Nature 2005, 434, 843–850. [Google Scholar] [CrossRef] [PubMed]
- Schneikert, J.; Behrens, J. The canonical Wnt signalling pathway and its APC partner in colon cancer development. Gut 2007, 56, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Novellasdemunt, L.; Antas, P.; Li, V.S. Targeting Wnt signaling in colorectal cancer. A review in the theme: Cell signaling: Proteins, pathways and mechanisms. Am. J. Physiol. Cell Physiol. 2015, 309, C511–C521. [Google Scholar] [CrossRef] [PubMed]
- Waisberg, J.; Saba, G.T. Wnt-/-β-catenin pathway signaling in human hepatocellular carcinoma. World J. Hepatol. 2015, 7, 2631–2635. [Google Scholar] [CrossRef] [PubMed]
- Chiurillo, M.A. Role of the Wnt/β-catenin pathway in gastric cancer: An in-depth literature review. World J. Exp. Med. 2015, 5, 84–102. [Google Scholar] [CrossRef] [PubMed]
- Pohl, S.G.; Brook, N.; Agostino, M.; Arfuso, F.; Kumar, A.P.; Dharmarajan, A. Wnt signaling in triple-negative breast cancer. Oncogenesis 2017, 6, e310. [Google Scholar] [CrossRef] [PubMed]
- Ng, O.H.; Erbilgin, Y.; Firtina, S.; Celkan, T.; Karakas, Z.; Aydogan, G.; Turkkan, E.; Yildirmak, Y.; Timur, C.; Zengin, E.; et al. Deregulated WNT signaling in childhood T-cell acute lymphoblastic leukemia. Blood Cancer J. 2014, 4, e192. [Google Scholar] [CrossRef] [PubMed]
- Molinolo, A.A.; Amornphimoltham, P.; Squarize, C.H.; Castilho, R.M.; Patel, V.; Gutkind, J.S. Dysregulated molecular networks in head and neck carcinogenesis. Oral Oncol. 2009, 45, 324–334. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Zeng, Q.; Yu, G.; Li, S.; Wang, C.Y. Wnt/β-catenin signaling inhibits death receptor-mediated apoptosis and promotes invasive growth of HNSCC. Cell Signal. 2006, 18, 679–687. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Chang, I.; Chen, Z.; Kang, M.; Wang, C.Y. Characterization of side populations in HNSCC: Highly invasive, chemoresistant and abnormal Wnt signaling. PLoS ONE 2010, 5, e11456. [Google Scholar] [CrossRef] [PubMed]
- Psyrri, A.; Kotoula, V.; Fountzilas, E.; Alexopoulou, Z.; Bobos, M.; Televantou, D.; Karayannopoulou, G.; Krikelis, D.; Markou, K.; Karasmanis, I.; et al. Prognostic significance of the Wnt pathway in squamous cell laryngeal cancer. Oral. Oncol. 2014, 50, 298–305. [Google Scholar] [CrossRef] [PubMed]
- Paluszczak, J.; Sarbak, J.; Kostrzewska-Poczekaj, M.; Kiwerska, K.; Jarmuz-Szymczak, M.; Grenman, R.; Mielcarek-Kuchta, D.; Baer-Dubowska, W. The negative regulators of Wnt pathway-DACH1, DKK1, and WIF1 are methylated in oral and oropharyngeal cancer and WIF1 methylation predicts shorter survival. Tumour Biol. 2015, 36, 2855–2861. [Google Scholar] [CrossRef] [PubMed]
- Schussel, J.L.; Kalinke, L.P.; Sassi, L.M.; de Oliveira, B.V.; Pedruzzi, P.A.; Olandoski, M.; Alvares, L.E.; Garlet, G.P.; Trevilatto, P.C. Expression and epigenetic regulation of DACT1 and DACT2 in oral squamous cell carcinoma. Cancer Biomark. 2015, 15, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wang, L.; Zhu, L.; Zhang, C.; Zhou, J. Secreted frizzledrelated protein 2 is epigenetically silenced and functions as a tumor suppressor in oral squamous cell carcinoma. Mol. Med. Rep. 2014, 10, 2293–2298. [Google Scholar] [PubMed]
- Honjo, Y.; Inohara, H.; Akahani, S.; Yoshii, T.; Takenaka, Y.; Yoshida, J.; Hattori, K.; Tomiyama, Y.; Raz, A.; Kubo, T. Expression of cytoplasmic galectin-3 as a prognostic marker in tongue carcinoma. Clin. Cancer Res. 2000, 6, 4635–4640. [Google Scholar] [PubMed]
- Wang, L.P.; Chen, S.W.; Zhuang, S.M.; Li, H.; Song, M. Galectin-3 accelerates the progression of oral tongue squamous cell carcinoma via a Wnt/β-catenin-dependent pathway. Pathol. Oncol. Res. 2013, 19, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.C.; Zhu, Y.T.; Chen, S.Y.; Tseng, S.C. Wnt signaling induces epithelial-mesenchymal transition with proliferation in ARPE-19 cells upon loss of contact inhibition. Lab. Investig. 2012, 92, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Katase, N.; Lefeuvre, M.; Tsujigiwa, H.; Fujii, M.; Ito, S.; Tamamura, R.; Buery, R.R.; Gunduz, M.; Nagatsuka, H. Knockdown of Dkk-3 decreases cancer cell migration and invasion independently of the Wnt pathways in oral squamous cell carcinomaderived cells. Oncol. Rep. 2013, 29, 1349–1355. [Google Scholar] [PubMed]
- Yang, B.; Du, Z.; Gao, Y.T.; Lou, C.; Zhang, S.G.; Bai, T.; Wang, Y.J.; Song, W.Q. Methylation of Dickkopf-3 as a prognostic factor in cirrhosis-related hepatocellular carcinoma. World J. Gastroenterol. 2010, 16, 755–763. [Google Scholar] [CrossRef] [PubMed]
- Veeck, J.; Wild, P.J.; Fuchs, T.; Schuffler, P.J.; Hartmann, A.; Knuchel, R.; Dahl, E. Prognostic relevance of Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter methylation in human breast cancer. BMC Cancer 2009, 9, 217. [Google Scholar] [CrossRef] [PubMed]
- Zenzmaier, C.; Untergasser, G.; Hermann, M.; Dirnhofer, S.; Sampson, N.; Berger, P. Dysregulation of Dkk-3 expression in benign and malignant prostatic tissue. Prostate 2008, 68, 540–547. [Google Scholar] [CrossRef] [PubMed]
- Katase, N.; Lefeuvre, M.; Gunduz, M.; Gunduz, E.; Beder, L.B.; Grenman, R.; Fujii, M.; Tamamura, R.; Tsujigiwa, H.; Nagatsuka, H. Absence of Dickkopf (Dkk)-3 protein expression is correlated with longer disease-free survival and lower incidence of metastasis in head and neck squamous cell carcinoma. Oncol. Lett. 2012, 3, 273–280. [Google Scholar] [CrossRef] [PubMed]
- Sasai, N.; Nakazawa, Y.; Haraguchi, T.; Sasai, Y. The neurotrophin-receptor-related protein NRH1 is essential for convergent extension movements. Nat. Cell Biol. 2004, 6, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.; Yamamoto, V.; Ortega, B.; Baltimore, D. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell 2004, 119, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Borchers, A.G.; Jolicoeur, C.; Rayburn, H.; Baker, J.C.; Tessier-Lavigne, M. PTK7/CCK-4 is a novel regulator of planar cell polarity in vertebrates. Nature 2004, 430, 93–98. [Google Scholar] [CrossRef] [PubMed]
- Nishita, M.; Yoo, S.K.; Nomachi, A.; Kani, S.; Sougawa, N.; Ohta, Y.; Takada, S.; Kikuchi, A.; Minami, Y. Filopodia formation mediated by receptor tyrosine kinase Ror2 is required for Wnt5a-induced cell migration. J. Cell Biol. 2006, 175, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Prgomet, Z.; Axelsson, L.; Lindberg, P.; Andersson, T. Migration and invasion of oral squamous carcinoma cells is promoted by WNT5A, a regulator of cancer progression. J. Oral Pathol. Med. 2015, 44, 776–784. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, A.; Iwai, S.; Morita, Y.; Niki-Yonekawa, A.; Hamada, M.; Yura, Y. Wnt5b promotes the cell motility essential for metastasis of oral squamous cell carcinoma through active Cdc42 and RhoA. Int. J. Oncol. 2014, 44, 59–68. [Google Scholar] [PubMed]
- Tran, F.H.; Zheng, J.J. Modulating the wnt signaling pathway with small molecules. Protein Sci. 2017, 26, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pan, S.; Hsieh, M.H.; Ng, N.; Sun, F.; Wang, T.; Kasibhatla, S.; Schuller, A.G.; Li, A.G.; Cheng, D.; et al. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc. Natl. Acad. Sci. USA 2013, 110, 20224–20229. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.J.; Lai, G.M.; Yeh, C.T.; Lai, M.T.; Shih, P.H.; Chao, W.J.; Whang-Peng, J.; Chuang, S.E.; Lai, T.Y. Honokiol eliminates human oral cancer stem-like cells accompanied with suppression of Wnt/ β -catenin signaling and apoptosis induction. Evid. Based Complement. Altern. Med. 2013, 2013, 146136. [Google Scholar] [CrossRef] [PubMed]
- Bain, V.E.; Gordon, J.; O’Neil, J.D.; Ramos, I.; Richie, E.R.; Manley, N.R. Tissue-specific roles for sonic hedgehog signaling in establishing thymus and parathyroid organ fate. Development 2016, 143, 4027–4037. [Google Scholar] [CrossRef] [PubMed]
- Strzyz, P. Adult stem cells: Hair stem cells born without a home. Nat. Rev. Mol. Cell Biol. 2016, 17, 133. [Google Scholar] [CrossRef] [PubMed]
- Faigle, R.; Song, H. Signaling mechanisms regulating adult neural stem cells and neurogenesis. Biochim. Biophys. Acta 2013, 1830, 2435–2448. [Google Scholar] [CrossRef] [PubMed]
- Rimkus, T.K.; Carpenter, R.L.; Qasem, S.; Chan, M.; Lo, H.W. Targeting the sonic hedgehog signaling pathway: Review of smoothened and GLI inhibitors. Cancers 2016, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Rubin, L.L.; de Sauvage, F.J. Targeting the Hedgehog pathway in cancer. Nat. Rev. Drug Discov. 2006, 5, 1026–1033. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.G.; Ye, L.; Ruge, F.; Sun, P.H.; Sanders, A.J.; Ji, K.; Lane, J.; Zhang, L.; Satherley, L.; Weeks, H.P.; et al. Expression of Sonic Hedgehog (SHH) in human lung cancer and the impact of YangZheng XiaoJi on SHH-mediated biological function of lung cancer cells and tumor growth. Anticancer Res. 2015, 35, 1321–1331. [Google Scholar] [PubMed]
- Noman, A.S.; Uddin, M.; Rahman, M.Z.; Nayeem, M.J.; Alam, S.S.; Khatun, Z.; Wahiduzzaman, M.; Sultana, A.; Rahman, M.L.; Ali, M.Y.; et al. Overexpression of sonic hedgehog in the triple negative breast cancer: Clinicopathological characteristics of high burden breast cancer patients from Bangladesh. Sci. Rep. 2016, 6, 18830. [Google Scholar] [CrossRef] [PubMed]
- Srinath, S.; Iyengar, A.R.; Mysorekar, V. Sonic hedgehog in oral squamous cell carcinoma: An immunohistochemical study. J. Oral Maxillofac. Pathol. 2016, 20, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.X.; Wang, S.; Zhao, H.; Liu, N.; Chen, D.; Sun, M.; Zheng, J.H. Sonic hedgehog signaling may promote invasion and metastasis of oral squamous cell carcinoma by activating MMP-9 and E-cadherin expression. Med. Oncol. 2014, 31, 41. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.F.; Chang, C.J.; Lin, C.P.; Chang, S.Y.; Chu, P.Y.; Tai, S.K.; Li, W.Y.; Chao, K.S.; Chen, Y.J. Expression of hedgehog signaling molecules as a prognostic indicator of oral squamous cell carcinoma. Head Neck 2012, 34, 1556–1561. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Wang, L.; Zuo, H.; Zhang, Z.; Chen, W.; Mao, L.; Zhang, P. HH/GLI signalling as a new therapeutic target for patients with oral squamous cell carcinoma. Oral Oncol. 2011, 47, 504–509. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli Buim, M.E.; Gurgel, C.A.; Goncalves Ramos, E.A.; Lourenco, S.V.; Soares, F.A. Activation of sonic hedgehog signaling in oral squamous cell carcinomas: A preliminary study. Hum. Pathol. 2011, 42, 1484–1490. [Google Scholar] [CrossRef] [PubMed]
- Honami, T.; Shimo, T.; Okui, T.; Kurio, N.; Hassan, N.M.; Iwamoto, M.; Sasaki, A. Sonic hedgehog signaling promotes growth of oral squamous cell carcinoma cells associated with bone destruction. Oral Oncol. 2012, 48, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.K.; Taipale, J.; Cooper, M.K.; Beachy, P.A. Inhibition of Hedgehog signaling by direct binding of cyclopamine to Smoothened. Genes Dev. 2002, 16, 2743–2748. [Google Scholar] [CrossRef] [PubMed]
- Mozet, C.; Stoehr, M.; Dimitrova, K.; Dietz, A.; Wichmann, G. Hedgehog targeting by cyclopamine suppresses head and neck squamous cell carcinoma and enhances chemotherapeutic effects. Anticancer Res. 2013, 33, 2415–2424. [Google Scholar] [PubMed]
- Gupta, S.; Takebe, N.; Lorusso, P. Targeting the Hedgehog pathway in cancer. Ther. Adv. Med. Oncol. 2010, 2, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Hann, C.L.; Laterra, J.; Yauch, R.L.; Callahan, C.A.; Fu, L.; Holcomb, T.; Stinson, J.; Gould, S.E.; Coleman, B.; et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N. Engl. J. Med. 2009, 361, 1173–1178. [Google Scholar] [CrossRef] [PubMed]
- Von Hoff, D.D.; LoRusso, P.M.; Rudin, C.M.; Reddy, J.C.; Yauch, R.L.; Tibes, R.; Weiss, G.J.; Borad, M.J.; Hann, C.L.; Brahmer, J.R.; et al. Inhibition of the hedgehog pathway in advanced basal-cell carcinoma. N. Engl. J. Med. 2009, 361, 1164–1172. [Google Scholar] [CrossRef] [PubMed]
- Scales, S.J.; de Sauvage, F.J. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol. Sci. 2009, 30, 303–312. [Google Scholar] [CrossRef] [PubMed]
- Stanton, B.Z.; Peng, L.F. Small-molecule modulators of the Sonic Hedgehog signaling pathway. Mol. Biosyst. 2010, 6, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Laterra, J.; Pomper, M.G. Hedgehog pathway inhibitor HhAntag691 is a potent inhibitor of ABCG2/BCRP and ABCB1/Pgp. Neoplasia 2009, 11, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.C.; Chao, K.S.; Liao, H.F.; Chen, Y.J. Targeting sonic hedgehog signaling by compounds and derivatives from natural products. Evid. Based Complement. Altern. Med. 2013, 2013, 748587. [Google Scholar] [CrossRef] [PubMed]
- Meulmeester, E.; Ten Dijke, P. The dynamic roles of TGF-β in cancer. J. Pathol. 2011, 223, 205–218. [Google Scholar] [CrossRef] [PubMed]
- Papageorgis, P.; Stylianopoulos, T. Role of TGFβ in regulation of the tumor microenvironment and drug delivery. Int. J. Oncol. 2015, 46, 933–943. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, S.; Bradshaw, A.D.; Gera, S.; Dewan, M.Z.; Xu, R. The TGF-β/Smad4 signaling pathway in pancreatic carcinogenesis and its clinical significance. J. Clin. Med. 2017, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Murata, M.; Yamaguchi, T.; Matsuzaki, K. TGF-β/Smad signaling during hepatic fibro-carcinogenesis. Int. J. Oncol. 2014, 45, 1363–1371. [Google Scholar] [CrossRef] [PubMed]
- Sivadas, V.P.; George, N.A.; Kattoor, J.; Kannan, S. Novel mutations and expression alterations in SMAD3/TGFBR2 genes in oral carcinoma correlate with poor prognosis. Genes. Chromosom. Cancer 2013, 52, 1042–1052. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.K.; Huang, A.H.; Cheng, P.H.; Yang, S.H.; Lin, L.M. Overexpression of Smad proteins, especially Smad7, in oral epithelial dysplasias. Clin. Oral Investig. 2013, 17, 921–932. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.K.; Yang, S.H.; Huang, A.H.; Hsue, S.S.; Lin, L.M. Aberrant expression in multiple components of the transforming growth factor-β1-induced Smad signaling pathway during 7,12-dimethylbenz[a]anthracene-induced hamster buccal-pouch squamous-cell carcinogenesis. Oral Oncol. 2011, 47, 262–267. [Google Scholar] [CrossRef] [PubMed]
- Mangone, F.R.; Walder, F.; Maistro, S.; Pasini, F.S.; Lehn, C.N.; Carvalho, M.B.; Brentani, M.M.; Snitcovsky, I.; Federico, M.H. Smad2 and Smad6 as predictors of overall survival in oral squamous cell carcinoma patients. Mol. Cancer 2010, 9, 106. [Google Scholar] [CrossRef] [PubMed]
- Park, I.; Son, H.K.; Che, Z.M.; Kim, J. A novel gain-of-function mutation of TGF-β receptor II promotes cancer progression via delayed receptor internalization in oral squamous cell carcinoma. Cancer Lett. 2012, 315, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Uehara, E.; Shiiba, M.; Shinozuka, K.; Saito, K.; Kouzu, Y.; Koike, H.; Kasamatsu, A.; Sakamoto, Y.; Ogawara, K.; Uzawa, K.; et al. Upregulated expression of ADAM12 is associated with progression of oral squamous cell carcinoma. Int. J. Oncol. 2012, 40, 1414–1422. [Google Scholar] [PubMed]
- Saito, D.; Kyakumoto, S.; Chosa, N.; Ibi, M.; Takahashi, N.; Okubo, N.; Sawada, S.; Ishisaki, A.; Kamo, M. Transforming growth factor-β1 induces epithelial-mesenchymal transition and integrin α3β1-mediated cell migration of HSC-4 human squamous cell carcinoma cells through Slug. J. Biochem. 2013, 153, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.G.; Song, J.Y. Therapeutic targeting of oncogenic transforming growth factor-β1 signaling by antisense oligonucleotides in oral squamous cell carcinoma. Oncol. Rep. 2012, 28, 539–544. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- MacDonagh, L.; Gray, S.G.; Finn, S.P.; Cuffe, S.; O’Byrne, K.J.; Barr, M.P. The emerging role of microRNAs in resistance to lung cancer treatments. Cancer Treat. Rev. 2015, 41, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Gorrini, C.; Harris, I.S.; Mak, T.W. Modulation of oxidative stress as an anticancer strategy. Nat. Rev. Drug Discov. 2013, 12, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Gandhi, V. ROS-activated anticancer prodrugs: A new strategy for tumor-specific damage. Ther. Deliv. 2012, 3, 823–833. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.C.; Tseng, C.N.; Yang, J.I.; Huang, H.W.; Fang, Y.; Tang, J.Y.; Chang, F.R.; Chang, H.W. Antiproliferation and induction of apoptosis in Ca9-22 oral cancer cells by ethanolic extract of Gracilaria tenuistipitata. Molecules 2012, 17, 10916–10927. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.H.; Farooqi, A.A.; Li, K.T.; Butt, G.; Tang, J.Y.; Wu, C.Y.; Cheng, Y.B.; Hou, M.F.; Chang, H.W. Methanolic extracts of Solieria robusta inhibits proliferation of oral cancer Ca9-22 cells via apoptosis and oxidative stress. Molecules 2014, 19, 18721–18732. [Google Scholar] [CrossRef] [PubMed]
- Yen, C.Y.; Chiu, C.C.; Haung, R.W.; Yeh, C.C.; Huang, K.J.; Chang, K.F.; Hseu, Y.C.; Chang, F.R.; Chang, H.W.; Wu, Y.C. Antiproliferative effects of goniothalamin on Ca9-22 oral cancer cells through apoptosis, DNA damage and ROS induction. Mutat. Res. 2012, 747, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Jeong, E.K.; Ju, M.K.; Jeon, H.M.; Kim, M.Y.; Kim, C.H.; Park, H.G.; Han, S.I.; Kang, H.S. Induction of metastasis, cancer stem cell phenotype, and oncogenic metabolism in cancer cells by ionizing radiation. Mol. Cancer 2017, 16, 10. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Zhu, Z.; Zhang, X.; Zhang, N.; Yao, Z. Idelalisib induces PUMA-dependent apoptosis in colon cancer cells. Oncotarget 2017, 8, 6102–6113. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Wang, H.; Li, W.; Zhong, J.; Yu, R.; Huang, X.; Wang, H.; Tan, Z.; Wang, J.; Zhang, Y. Pazopanib, a novel multi-kinase inhibitor, shows potent antitumor activity in colon cancer through PUMA-mediated apoptosis. Oncotarget 2017, 8, 3289–3303. [Google Scholar] [CrossRef] [PubMed]
- Lu, W.C.; Kao, S.Y.; Yang, C.C.; Tu, H.F.; Wu, C.H.; Chang, K.W.; Lin, S.C. EGF up-regulates miR-31 through the C/EBPβ signal cascade in oral carcinoma. PLoS ONE 2014, 9, e108049. [Google Scholar] [CrossRef] [PubMed]
- Tseng, S.H.; Yang, C.C.; Yu, E.H.; Chang, C.; Lee, Y.S.; Liu, C.J.; Chang, K.W.; Lin, S.C. K14-EGFP-miR-31 transgenic mice have high susceptibility to chemical-induced squamous cell tumorigenesis that is associating with Ku80 repression. Int. J. Cancer 2015, 136, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Ren, Y.; Liu, A.; Han, L.; Zhang, K.; Li, S.; Li, P.; Li, P.; Kang, C.; Wang, X.; et al. STAT3 inhibitor WP1066 attenuates miRNA-21 to suppress human oral squamous cell carcinoma growth in vitro and in vivo. Oncol. Rep. 2014, 31, 2173–2180. [Google Scholar] [CrossRef] [PubMed]
- Jiang, F.; Zhao, W.; Zhou, L.; Zhang, L.; Liu, Z.; Yu, D. miR-222 regulates the cell biological behavior of oral squamous cell carcinoma by targeting PUMA. Oncol. Rep. 2014, 31, 1255–1262. [Google Scholar] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acid. Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Wang, Z.; Wang, Y.; Gu, W. MiR-338 suppresses the growth and metastasis of OSCC cells by targeting NRP1. Mol. Cell. Biochem. 2015, 398, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Sun, L.; Chen, W.; Liu, B.; Wang, Y.; Fan, S.; Li, Y.; Li, J. miR-639 regulates transforming growth factor β-induced epithelial-mesenchymal transition in human tongue cancer cells by targeting FOXC1. Cancer Sci. 2014, 105, 1288–1298. [Google Scholar] [CrossRef] [PubMed]
- Song, K.B.; Liu, W.J.; Jia, S.S. miR-219 inhibits the growth and metastasis of TSCC cells by targeting PRKCI. Int. J. Clin. Exp. Med. 2014, 7, 2957–2965. [Google Scholar] [PubMed]
- Shiah, S.G.; Hsiao, J.R.; Chang, W.M.; Chen, Y.W.; Jin, Y.T.; Wong, T.Y.; Huang, J.S.; Tsai, S.T.; Hsu, Y.M.; Chou, S.T.; et al. Downregulated miR329 and miR410 promote the proliferation and invasion of oral squamous cell carcinoma by targeting Wnt-7b. Cancer Res. 2014, 74, 7560–7572. [Google Scholar] [CrossRef] [PubMed]
- Kariya, A.; Furusawa, Y.; Yunoki, T.; Kondo, T.; Tabuchi, Y. A microRNA-27a mimic sensitizes human oral squamous cell carcinoma HSC-4 cells to hyperthermia through downregulation of Hsp110 and Hsp90. Int. J. Mol. Med. 2014, 34, 334–340. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Shivananda, S.; Gopinath, K.S.; Kumar, A. MicroRNA-125a reduces proliferation and invasion of oral squamous cell carcinoma cells by targeting estrogen-related receptor α: Implications for cancer therapeutics. J. Biol. Chem. 2014, 289, 32276–32290. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Wu, H.; Ling, T. Suppressive effect of microRNA-126 on oral squamous cell carcinoma in vitro. Mol. Med. Rep. 2014, 10, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Yen, Y.C.; Shiah, S.G.; Chu, H.C.; Hsu, Y.M.; Hsiao, J.R.; Chang, J.Y.; Hung, W.C.; Liao, C.T.; Cheng, A.J.; Lu, Y.C.; et al. Reciprocal regulation of microRNA-99a and insulin-like growth factor I receptor signaling in oral squamous cell carcinoma cells. Mol. Cancer 2014, 13, 6. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.C.; Chan, S.H.; Jang, T.H.; Chang, J.W.; Ko, Y.C.; Yen, T.C.; Chiang, S.L.; Chiang, W.F.; Shieh, T.Y.; Liao, C.T.; et al. miRNA-491–5p and GIT1 serve as modulators and biomarkers for oral squamous cell carcinoma invasion and metastasis. Cancer Res. 2014, 74, 751–764. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Liu, K.; Wu, Y.; Fan, J.; Chen, J.; Li, C.; Yang, Q.; Wang, Z. MicroRNA-9 inhibits the proliferation of oral squamous cell carcinoma cells by suppressing expression of CXCR4 via the Wnt/β-catenin signaling pathway. Oncogene 2014, 33, 5017–5027. [Google Scholar] [CrossRef] [PubMed]
- Kawakita, A.; Yanamoto, S.; Yamada, S.; Naruse, T.; Takahashi, H.; Kawasaki, G.; Umeda, M. MicroRNA-21 promotes oral cancer invasion via the Wnt/β-catenin pathway by targeting DKK2. Pathol. Oncol. Res. 2014, 20, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Wilmink, G.J.; Roth, C.L.; Ibey, B.L.; Ketchum, N.; Bernhard, J.; Cerna, C.Z.; Roach, W.P. Identification of microRNAs associated with hyperthermia-induced cellular stress response. Cell Stress Chaperones 2010, 15, 1027–1038. [Google Scholar] [CrossRef] [PubMed]
- Bernatchez, G.; Giroux, V.; Lassalle, T.; Carpentier, A.C.; Rivard, N.; Carrier, J.C. ERRα metabolic nuclear receptor controls growth of colon cancer cells. Carcinogenesis 2013, 34, 2253–2261. [Google Scholar] [CrossRef] [PubMed]
- Cavallini, A.; Notarnicola, M.; Giannini, R.; Montemurro, S.; Lorusso, D.; Visconti, A.; Minervini, F.; Caruso, M.G. Oestrogen receptor-related receptor α (ERRα) and oestrogen receptors (ERα and ERβ) exhibit different gene expression in human colorectal tumour progression. Eur. J. Cancer 2005, 41, 1487–1494. [Google Scholar] [CrossRef] [PubMed]
- Nikolic, I.; Plate, K.H.; Schmidt, M.H. EGFL7 meets miRNA-126: An angiogenesis alliance. J. Angiogenes Res. 2010, 2, 9. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Wang, L.; Zhu, L.; Zhang, C.; Zhou, J. Curcumin inhibits oral squamous cell carcinoma SCC-9 cells proliferation by regulating miR-9 expression. Biochem. Biophys. Res. Commun. 2014, 454, 576–580. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Li, K.T.; Fayyaz, S.; Chang, Y.T.; Ismail, M.; Liaw, C.C.; Yuan, S.S.; Tang, J.Y.; Chang, H.W. Anticancer drugs for the modulation of endoplasmic reticulum stress and oxidative stress. Tumour Biol. 2015, 36, 5743–5752. [Google Scholar] [CrossRef] [PubMed]
- Edagawa, M.; Kawauchi, J.; Hirata, M.; Goshima, H.; Inoue, M.; Okamoto, T.; Murakami, A.; Maehara, Y.; Kitajima, S. Role of activating transcription factor 3 (ATF3) in endoplasmic reticulum (ER) stress-induced sensitization of p53-deficient human colon cancer cells to tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL)-mediated apoptosis through up-regulation of death receptor 5 (DR5) by zerumbone and celecoxib. J. Biol. Chem. 2014, 289, 21544–21561. [Google Scholar] [PubMed]
- Kim, J.; Yun, M.; Kim, E.O.; Jung, D.B.; Won, G.; Kim, B.; Jung, J.H.; Kim, S.H. Decursin enhances TRAIL-induced apoptosis through oxidative stress mediated-endoplasmic reticulum stress signalling in non-small cell lung cancers. Br. J. Pharmacol. 2016, 173, 1033–1044. [Google Scholar] [CrossRef] [PubMed]
- Tiwary, R.; Yu, W.; Li, J.; Park, S.K.; Sanders, B.G.; Kline, K. Role of endoplasmic reticulum stress in α-TEA mediated TRAIL/DR5 death receptor dependent apoptosis. PLoS ONE 2010, 5, e11865. [Google Scholar] [CrossRef] [PubMed]
- Martin-Perez, R.; Niwa, M.; Lopez-Rivas, A. ER stress sensitizes cells to TRAIL through down-regulation of FLIP and Mcl-1 and PERK-dependent up-regulation of TRAIL-R2. Apoptosis 2012, 17, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Wang, Y.; Li, X.; Chen, Z.; Li, X.; Wang, H.; Ni, M.; Li, J. Molecular mechanism of ER stress-induced gene expression of tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) in macrophages. FEBS J. 2015, 282, 2361–2378. [Google Scholar] [CrossRef] [PubMed]
- Zlotorynski, E. Apoptosis. DR5 unfolds ER stress. Nat. Rev. Mol. Cell Biol. 2014, 15, 498–499. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Hamao, A.; Tanaka, A.; Kitada, M.; Suzuki, S.; Kusama, K.; Sakashita, H. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL/APO2L) and its receptors expression in human squamous cell carcinoma of the oral cavity. Oncol. Rep. 2003, 10, 1113–1119. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Gao, M.; Wang, J.; Kong, Q.; Hua, H.; Luo, T.; Jiang, Y. Inhibition of eIF2α dephosphorylation enhances TRAIL-induced apoptosis in hepatoma cells. Cell Death Dis. 2014, 5, e1060. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Fan, C.; Yang, Y.; Di, S.; Hu, W.; Li, T.; Zhu, Y.; Han, J.; Xin, Z.; Wu, G.; et al. Thapsigargin sensitizes human esophageal cancer to TRAIL-induced apoptosis via AMPK activation. Sci. Rep. 2016, 6, 35196. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.I.; Wang, R.Y.; Lin, J.J.; Su, J.H.; Chiu, C.C.; Chen, J.C.; Chen, J.Y.; Wu, Y.J. Proteomic profiling of the 11-dehydrosinulariolide-treated oral carcinoma cells Ca9-22: Effects on the cell apoptosis through mitochondrial-related and ER stress pathway. J. Proteom. 2012, 75, 5578–5589. [Google Scholar] [CrossRef] [PubMed]
- Sidhu, A.; Miller, J.R.; Tripathi, A.; Garshott, D.M.; Brownell, A.L.; Chiego, D.J.; Arevang, C.; Zeng, Q.; Jackson, L.C.; Bechler, S.A.; et al. Borrelidin induces the unfolded protein response in oral cancer cells and Chop-dependent apoptosis. ACS Med. Chem. Lett. 2015, 6, 1122–1127. [Google Scholar] [CrossRef] [PubMed]
- El Jamal, S.M.; Taylor, E.B.; Abd Elmageed, Z.Y.; Alamodi, A.A.; Selimovic, D.; Alkhateeb, A.; Hannig, M.; Hassan, S.Y.; Santourlidis, S.; Friedlander, P.L.; et al. Interferon γ-induced apoptosis of head and neck squamous cell carcinoma is connected to indoleamine-2,3-dioxygenase via mitochondrial and ER stress-associated pathways. Cell Div. 2016, 11, 11. [Google Scholar] [CrossRef] [PubMed]
- Utaipan, T.; Athipornchai, A.; Suksamrarn, A.; Chunsrivirot, S.; Chunglok, W. Isomahanine induces endoplasmic reticulum stress and simultaneously triggers p38 MAPK-mediated apoptosis and autophagy in multidrug-resistant human oral squamous cell carcinoma cells. Oncol. Rep. 2017, 37, 1243–1252. [Google Scholar] [PubMed]
- Su, C.H.; Kuo, C.L.; Lu, K.W.; Yu, F.S.; Ma, Y.S.; Yang, J.L.; Chu, Y.L.; Chueh, F.S.; Liu, K.C.; Chung, J.G. Fisetin-induced apoptosis of human oral cancer SCC-4 cells through reactive oxygen species production, endoplasmic reticulum stress, caspase-, and mitochondria-dependent signaling pathways. Environ. Toxicol. 2017, 32, 1725–1741. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, Y.; Wang, L.; Yang, H.; Wang, Q.; Qi, H.; Li, S.; Zhou, P.; Liang, P.; Wang, Q.; et al. microRNA response elements-regulated TRAIL expression shows specific survival-suppressing activity on bladder cancer. J Exp. Clin. Cancer Res. 2013, 32, 10. [Google Scholar] [CrossRef] [PubMed]
- Catto, J.W.; Alcaraz, A.; Bjartell, A.S.; De Vere White, R.; Evans, C.P.; Fussel, S.; Hamdy, F.C.; Kallioniemi, O.; Mengual, L.; Schlomm, T.; et al. MicroRNA in prostate, bladder, and kidney cancer: A systematic review. Eur. Urol. 2011, 59, 671–681. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, L.; Li, C.; Zhang, Z.; Yang, G.; Zhang, W. Tumor-targeting TRAIL expression mediated by miRNA response elements suppressed growth of uveal melanoma cells. Mol. Oncol. 2013, 7, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Huo, W.; Jin, N.; Fan, L.; Wang, W. MiRNA regulation of TRAIL expression exerts selective cytotoxicity to prostate carcinoma cells. Mol. Cell. Biochem. 2014, 388, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Zhang, F.; Fan, Q.; Li, X.; Zhou, K. Breast cancer-specific TRAIL expression mediated by miRNA response elements of let-7 and miR-122. Neoplasma 2014, 61, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.; Jeon, Y.J.; Lagana, A.; Middleton, J.; Secchiero, P.; Garofalo, M.; Croce, C.M. MicroRNA-148a reduces tumorigenesis and increases TRAIL-induced apoptosis in NSCLC. Proc. Natl. Acad. Sci. USA 2015, 112, 8650–8655. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; Yaylim, I.; Ozkan, N.E.; Zaman, F.; Halim, T.A.; Chang, H.W. Restoring TRAIL mediated signaling in ovarian cancer cells. Arch. Immunol. Ther. Exp. 2014, 62, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, A.A.; De Rosa, G. TRAIL and microRNAs in the treatment of prostate cancer: Therapeutic potential and role of nanotechnology. Appl. Microbiol. Biotechnol. 2013, 97, 8849–8857. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Shao, N.; Ji, C. Targeting microRNAs to modulate TRAIL-induced apoptosis of cancer cells. Cancer Gene Ther. 2013, 20, 33–37. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhou, Q.; Tan, S. Targeting miRNAs associated with surface expression of death receptors to modulate TRAIL resistance in breast cancer. Cancer Lett. 2016, 383, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Garofalo, M.; Condorelli, G.L.; Croce, C.M.; Condorelli, G. MicroRNAs as regulators of death receptors signaling. Cell Death Differ. 2010, 17, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Razumilava, N.; Bronk, S.F.; Smoot, R.L.; Fingas, C.D.; Werneburg, N.W.; Roberts, L.R.; Mott, J.L. miR-25 targets TNF-related apoptosis inducing ligand (TRAIL) death receptor-4 and promotes apoptosis resistance in cholangiocarcinoma. Hepatology 2012, 55, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Shin, E.A.; Sohn, E.J.; Won, G.; Choi, J.U.; Jeong, M.; Kim, B.; Kim, M.J.; Kim, S.H. Upregulation of microRNA135a-3p and death receptor 5 plays a critical role in Tanshinone I sensitized prostate cancer cells to TRAIL induced apoptosis. Oncotarget 2014, 5, 5624–5636. [Google Scholar] [CrossRef] [PubMed]
- Dennler, S.; Andre, J.; Alexaki, I.; Li, A.; Magnaldo, T.; ten Dijke, P.; Wang, X.J.; Verrecchia, F.; Mauviel, A. Induction of sonic hedgehog mediators by transforming growth factor-β: Smad3-dependent activation of Gli2 and Gli1 expression in vitro and in vivo. Cancer Res. 2007, 67, 6981–6986. [Google Scholar] [CrossRef] [PubMed]
- Javelaud, D.; Pierrat, M.J.; Mauviel, A. Crosstalk between TGF-β and hedgehog signaling in cancer. FEBS Lett. 2012, 586, 2016–2025. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Deng, W.; Nail, C.D.; Bailey, S.K.; Kraus, M.H.; Ruppert, J.M.; Lobo-Ruppert, S.M. Snail induction is an early response to Gli1 that determines the efficiency of epithelial transformation. Oncogene 2006, 25, 609–621. [Google Scholar] [CrossRef] [PubMed]
- Aval, S.F.; Lotfi, H.; Sheervalilou, R.; Zarghami, N. Tuning of major signaling networks (TGF-β, Wnt, Notch and Hedgehog) by miRNAs in human stem cells commitment to different lineages: Possible clinical application. Biomed. Pharmacother. 2017, 91, 849–860. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhou, S.; Mao, L.; Zhang, H.; Sun, D.; Zhang, J.; Li, J.; Tang, J.H. Crosstalk between TGF-β signaling and miRNAs in breast cancer metastasis. Tumour Biol. 2016, 37, 10011–10019. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Zhang, Y.; Zhang, L.; Huang, F.; Li, J.; Wang, S. MicroRNAs, TGF-β signaling, and the inflammatory microenvironment in cancer. Tumour Biol. 2016, 37, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Onyido, E.K.; Sweeney, E.; Nateri, A.S. Wnt-signalling pathways and microRNAs network in carcinogenesis: Experimental and bioinformatics approaches. Mol. Cancer 2016, 15, 56. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhang, X.; Feng, X.; Fan, X.; Jin, Z. The crosstalk between microRNAs and the Wnt/β-catenin signaling pathway in cancer. Oncotarget 2017, 8, 14089–14106. [Google Scholar] [CrossRef] [PubMed]
- Rahmani, F.; Avan, A.; Hashemy, S.I.; Hassanian, S.M. Role of Wnt/β-catenin signaling regulatory microRNAs in the pathogenesis of colorectal cancer. J. Cell. Physiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Song, J.L.; Nigam, P.; Tektas, S.S.; Selva, E. MicroRNA regulation of Wnt signaling pathways in development and disease. Cell Signal. 2015, 27, 1380–1391. [Google Scholar] [CrossRef] [PubMed]
- Hyun, J.; Jung, Y. MicroRNAs in liver fibrosis: Focusing on the interaction with hedgehog signaling. World J. Gastroenterol. 2016, 22, 6652–6662. [Google Scholar] [CrossRef] [PubMed]

| Drugs/Natural Products | Gene Expression | OSCC Cell Lines | References |
|---|---|---|---|
| Smilax china L. extract (SCE) | 1. enhances DR5 2. reduces pERK | MC3 | [13] |
| β-Phenylethyl isothiocyanate (PEITC) | 1. enhances DR5 | HN22 | [14] |
| 2-deoxy-d-glucose | 1. enhances DR5 when combined with TRAIL | KB | [15] |
| Suberoylanilide hydroxamic acid (SAHA) | 1. enhances DR4, DR5, Fas, and the Fas ligand | Ca9-22, SAS | [16] |
| Esculetin | 1. enhances DR5 | SAS | [17] |
| S-1 (fluoropyrimidine anti-oral cancer agent) | 1. reduces tumor growth of OSCC-xenografting mice when combined with TRAIL | HSC2-xenografting mice | [18] |
| miRNAs | Target Genes | OSCC Cell Lines | References |
|---|---|---|---|
| Oncogenic miRNAs | |||
| miR-31 | Akt | HSC-3, OECM-1, SAS | [98] |
| miR-21 | STAT3 | TCA8113, TSCCA | [100] |
| miR-222 | PUMA | TCA8113, UM1 | [101] |
| Tumor suppressor miRNAs | |||
| miR-338 | NP-1 | TCA8113, SCC15 | [103] |
| miR-639 | FOXC1 | CAL 27, SCC9 | [104] |
| miR-219 | PRKCI | CAL 27, SCC15 | [105] |
| miR-410 | WNT7B | DOK, FaDu, OC-3, OEC-M1, SCC4, SCC9, SCC15, SCC25, Tw2.6, YD-15 | [106] |
| miR-27a | Hsp90, Hsp110 | HSC-4 | [107] |
| miR-125a | ERRα | SCC084, SCC131 | [108] |
| miR-126 | EGFL7 | OSCC-15 | [109] |
| miR-99a | IGF1R | CGHNC9, OC-3, OEC-M1, TW2.6, FaDu, KB, SCC4, SCC15, SCC9, SCC25, UT-MUC-1, YD-15, DOK, Tu183, UMSCC1, HSC3 | [110] |
| miR-491-5p | GIT1 | CGHNC9, SAS, SCC25, OECM-1, OC-3 | [111] |
| miR-9 | CXCR4 | TCA8113, SCC9 | [112] |
| miR-21 | DKK2 | SCC25 | [113] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farooqi, A.A.; Shu, C.-W.; Huang, H.-W.; Wang, H.-R.; Chang, Y.-T.; Fayyaz, S.; Yuan, S.-S.F.; Tang, J.-Y.; Chang, H.-W. TRAIL, Wnt, Sonic Hedgehog, TGFβ, and miRNA Signalings Are Potential Targets for Oral Cancer Therapy. Int. J. Mol. Sci. 2017, 18, 1523. https://doi.org/10.3390/ijms18071523
Farooqi AA, Shu C-W, Huang H-W, Wang H-R, Chang Y-T, Fayyaz S, Yuan S-SF, Tang J-Y, Chang H-W. TRAIL, Wnt, Sonic Hedgehog, TGFβ, and miRNA Signalings Are Potential Targets for Oral Cancer Therapy. International Journal of Molecular Sciences. 2017; 18(7):1523. https://doi.org/10.3390/ijms18071523
Chicago/Turabian StyleFarooqi, Ammad Ahmad, Chih-Wen Shu, Hurng-Wern Huang, Hui-Ru Wang, Yung-Ting Chang, Sundas Fayyaz, Shyng-Shiou F. Yuan, Jen-Yang Tang, and Hsueh-Wei Chang. 2017. "TRAIL, Wnt, Sonic Hedgehog, TGFβ, and miRNA Signalings Are Potential Targets for Oral Cancer Therapy" International Journal of Molecular Sciences 18, no. 7: 1523. https://doi.org/10.3390/ijms18071523
APA StyleFarooqi, A. A., Shu, C. -W., Huang, H. -W., Wang, H. -R., Chang, Y. -T., Fayyaz, S., Yuan, S. -S. F., Tang, J. -Y., & Chang, H. -W. (2017). TRAIL, Wnt, Sonic Hedgehog, TGFβ, and miRNA Signalings Are Potential Targets for Oral Cancer Therapy. International Journal of Molecular Sciences, 18(7), 1523. https://doi.org/10.3390/ijms18071523