Leptin Receptor Gene Variant rs11804091 Is Associated with BMI and Insulin Resistance in Spanish Female Obese Children: A Case-Control Study
Abstract
:1. Introduction
2. Results
2.1. General Characteristics of the Population
2.2. Association of LEPR SNPs with Obesity
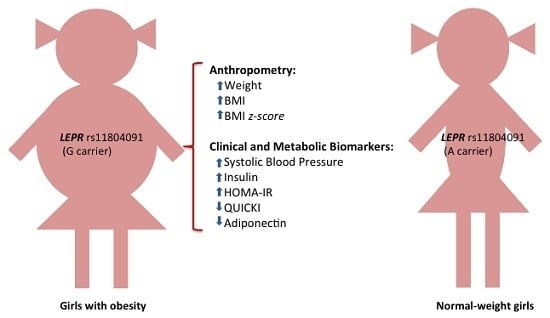
2.3. Association of SNP rs11804091 with Obesity-Related Traits
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Anthropometric and Biochemical Measurements
4.3. Inflammation and Cardiovascular Risk Biomarkers
4.4. DNA Isolation and Genotyping
4.5. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| Apo | Apolipoprotein |
| AST | Aspartate transaminase |
| BBB | Blood–brain barrier |
| BMI | Body mass index |
| BP | Blood pressure |
| C-RP | C-Reactive protein |
| CVD | Cardiovascular disease |
| ERK | Extracellular signal-regulated kinase |
| GGT | Gamma-glutamyl transpeptidase |
| HDL-c | High-density lipoprotein-cholesterol |
| HOMA-IR | Homeostatic model assessment of insulin resistance |
| IL | Interleukine |
| JAK | Janus kinase |
| LD | Linkage disequilibrium |
| LDL-c | Low-density lipoprotein-cholesterol |
| LEPR | Leptin receptor |
| MAF | Minor allele frequency |
| MMP-9 | Metalloproteinase-9 |
| MPO | Myeloperoxidase |
| PAI-1 | Plasminogen activator inhibitor |
| PI3K | Phosphatidylinositol 3′-kinase |
| POMC | Pro-opiomelanocortin |
| QUICKI | Quantitative insulin sensitivity check index |
| sICAM-1 | Soluble intracellular adhesion molecule-1 |
| SNP | Single nucleotide polymorphisms |
| TAG | Triacylglycerols |
| TNF-α | Tumour necrosis factor alpha |
| WC | Waist circumference |
| z-BMI | BMI z-Score |
References
- Chesi, A.; Grant, S. The genetics of pediatric obesity. Trends Endocrinol. Metab. 2015, 26, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Manco, M.; Dallapiccola, B. Genetics of pediatric obesity. Pediatrics 2012, 130, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Fleming, T.; Robinson, M.; Thomson, B.; Graetz, N.; Margono, C.; Mullany, E.C.; Biryukov, S.; Abbafati, C.; Abera, S.F.; et al. Global, regional, and national prevalence of overweight and obesity in children and adults during 1980–2013: A systematic analysis for the Global Burden of Disease Study 2013. Lancet 2014, 384, 766–781. [Google Scholar] [CrossRef]
- Fernandez, J.R.; Klimentidis, Y.C.; Dulin-Keita, A.; Casazza, K. Genetic influences in childhood obesity: Recent progress and recommendations for experimental designs. Int. J. Obes. 2012, 36, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Crujeiras, A.B.; Carreira, M.C.; Cabia, B.; Andrade, S.; Amil, M.; Casanueva, F.F. Leptin resistance in obesity: An epigenetics landscape. Life Sci. 2015, 140, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Hirako, S.; Takenoya, F.; Kageyama, H.; Okabe, M.; Shioda, S. Leptin and its receptors. J. Chem. Neuroanat. 2014, 61–62, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Gill, R.; Cheung, Y.H.; Shen, Y.; Lanzano, P.; Mirza, N.M.; Ten, S.; Maclaren, N.K.; Motaghedi, R.; Han, J.C.; Yanovski, J.A.; et al. Whole-exome sequencing identifies novel LEPR mutation in individuals with severe early onset obesity. Obesity 2014, 22, 576–584. [Google Scholar] [CrossRef] [PubMed]
- Leggio, A.; Catalano, S.; de Marco, R.; Barone, I.; Andò, S.; Liguori, A. Therapeutic potential of leptin receptor modulators. Eur. J. Med. Chem. 2014, 78, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Paz-Fhilo, G.; Mastronardi, C.; Wong, M.L.; Licinio, J. Leptin therapy, insulin sensitivity, and glucose homeostasis. Indian J. Endocr. Metab. 2012, 16, S549–S555. [Google Scholar] [CrossRef] [PubMed]
- Sainz, N.; Barrenetxe, J.; Moreno-Aliaga, M.J.; Martínez, J.A. Leptin resistance and diet-induced obesity: Central and peripheral actions of leptin. Metabolism 2015, 64, 35–46. [Google Scholar]
- Dubern, B.; Clement, K. Leptin and leptin receptor-related monogenic obesity. Biochimie 2012, 94, 2111–2115. [Google Scholar] [CrossRef] [PubMed]
- Farooqi, I.S.; Wangensteen, T.; Collins, S.; Kimber, W.; Matarese, G.; Keogh, J.M.; Lank, E.; Bottomley, B.; Lopez-Fernandez, J.; Ferraz-Amaro, I. Clinical and molecular genetic spectrum of congenital deficiency of the leptin receptor. N. Engl. J. Med. 2007, 18, 237–247. [Google Scholar] [CrossRef] [PubMed]
- Wheeler, E.; Huang, N.; Bochukova, E.G.; Keogh, J.M.; Lindsay, S.; Garg, S.; Henning, E.; Blackburn, H.; Loos, R.J.; Wareham, N.J.; et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet. 2013, 45, 513–517. [Google Scholar] [CrossRef] [PubMed]
- Heo, M.; Leibel, R.L.; Fontaine, K.R.; Boyer, B.B.; Chung, W.K.; Koulu, M.; Karvonen, M.K.; Pesonen, U.; Rissanen, A.; Laakso, M.; et al. A meta-analytic investigation of linkage and association of common leptin receptor (LEPR) polymorphisms with body mass index and waist circumference. Int. J. Obes. 2002, 26, 640–646. [Google Scholar] [CrossRef] [PubMed]
- Kilpeläinen, T.O.; Carli, J.F.; Skowronski, A.A.; Sun, Q.; Kriebel, J.; Feitosa, M.F.; Hedman, Å.K.; Drong, A.W.; Hayes, J.E.; Zhao, J.; et al. Genome-wide meta-analysis uncovers novel loci influencing circulating leptin levels. Nat. Commun. 2016, 7, 10494. [Google Scholar] [CrossRef] [PubMed]
- Portolés, O.; Sorlí, J.V.; Francés, F.; Coltell, O.; González, J.I.; Sáiz, C.; Corella, D. Effect of genetic variation in the leptin gene promoter and the leptin receptor gene on obesity risk in a population-based case-control study n Spain. Eur. J. Epidemiol. 2006, 21, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.S.; Heard-Costa, N.; Cupples, L.A.; Dupuis, J.; Vasan, R.S.; Atwood, L.D. Genome-wide association to body mass index and waist circumference: The Framingham Heart Study 100K project. BMC Med. Genet. 2007, 8, S18. [Google Scholar] [CrossRef] [PubMed]
- Gallicchio, L.; Chang, H.H.; Christo, D.K.; Thuita, L.; Huang, H.Y.; Strickland, P.; Ruczinski, I.; Clipp, S.; Helzlsouer, K.J. Single nucleotide polymorphisms in obesity-related genes and all-cause and cause-specific mortality: A prospective cohort study. BMC Med. Genet. 2009, 10, 103. [Google Scholar] [CrossRef] [PubMed]
- Furusawa, T.; Naka, I.; Yamauchi, T.; Natsuhara, K.; Kimura, R.; Nakazawa, M.; Ishida, T.; Inaoka, T.; Matsumura, Y.; Ataka, Y.; et al. The Q223R polymorphism in LEPR is associated with obesity in Pacific Islanders. Hum. Genet. 2010, 127, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Bender, N.; Allemann, N.; Marek, D.; Vollenweider, P.; Waeber, G.; Mooser, V.; Egger, M.; Bochud, M. Association between variants of the leptin receptor gene (LEPR) and overweight: A systematic review and an analysis of the CoLaus study. PLoS ONE 2011, 6, e26157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Vimaleswaran, K.S.; Angquist, L.; Hansen, R.D.; van der A, D.L.; Holst, C.; Tjønneland, A.; Overvad, K.; Jakobsen, M.U.; Boeing, H.; et al. Genetic polymorphisms in the hypothalamic pathway in relation to subsequent weight change—The DiOGenes study. PLoS ONE 2011, 6, e17436. [Google Scholar] [CrossRef] [PubMed]
- Boumaiza, I.; Omezzine, A.; Rejeb, J.; Rebhi, L.; Ouedrani, A.; Ben Rejeb, N.; Nabli, N.; Ben Abdelaziz, A.; Bouslama, A. Relationship between leptin G2548A and leptin receptor Q223R gene polymorphisms and obesity and metabolic syndrome risk in Tunisian volunteers. Genet. Test. Mol. Biomark. 2012, 16, 726–733. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Han, S.; Cao, X.; Zhu, C.; Wang, X.; Guo, X. Genetic polymorphisms in adipokine genes and the risk of obesity: A systematic review and meta-analysis. Obesity (Silver Spring) 2012, 20, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Komşu-Ornek, Z.; Demirel, F.; Dursun, A.; Ermiş, B.; Pişkin, E.; Bideci, A. Leptin receptor gene Gln223Arg polymorphism is not associated with obesity and metabolic syndrome in Turkish children. Turk. J. Pediatr. 2012, 54, 20–24. [Google Scholar] [PubMed]
- Pyrzak, B.; Wisniewska, A.; Kucharska, A. No association of LEPR GLN 223ARG polymorphism with leptin, obesity or metabolic disturbances in children. Eur. J. Med. Res. 2009, 14SIV, 201–204. [Google Scholar] [CrossRef]
- Angel-Chavez, L.I.; Tene-Pérez, C.E.; Castro, E. Leptin receptor gene K656N polymorphism is associated with low body fat levels and elevated high-density cholesterol levels in Mexican children and adolescents. Endocr. Res. 2012, 37, 124–134. [Google Scholar] [CrossRef] [PubMed]
- Labayen, I.; Ruiz, J.R.; Moreno, L.A.; Ortega, F.B.; Beghin, L.; DeHenauw, S.; Benito, P.J.; Diaz, L.E.; Ferrari, M.; Moschonis, G.; et al. The effect of ponderal index at birth on the relationships between common LEP and LEPR polymorphisms and adiposity in adolescents. Obesity (Silver Spring, Md) 2011, 19, 2038–2045. [Google Scholar] [CrossRef] [PubMed]
- Tabassum, R.; Mahendran, Y.; Dwivedi, O.P.; Chauhan, G.; Ghosh, S.; Marwaha, R.K.; Tandon, N.; Bharadwaj, D. Common variants of IL6, LEPR, and PBEF1 are associated with obesity in Indian children. Diabetes 2012, 61, 626–631. [Google Scholar] [CrossRef] [PubMed]
- Riestra, P.; García-Anguita, A.; Schoppen, S.; López-Simón, L.; de Oya, M.; Garcés, C. Sex-specific association between leptin receptor polymorphisms and leptin levels and BMI in healthy adolescents. Acta Paediatr. 2010, 99, 1527–1530. [Google Scholar] [CrossRef] [PubMed]
- Gajewska, J.; Kuryłowicz, A.; Ambroszkiewicz, J.; Mierzejewska, E.; Chełchowska, M.; Szamotulska, K.; Weker, H.; Puzianowska-Kuźnicka, M. ADIPOQ -11377C>G Polymorphism Increases the Risk of Adipokine Abnormalities and Child Obesity Regardless of Dietary Intake. J. Pediatr. Gastroenterol. Nutr. 2016, 62, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.M.; Wang, J.; Fan, J.J.; Ng, T.K.; Sun, D.J.; Guo, X.; Teng, Y.; Li, Y.B. Variations in the Obesity Gene “LEPR” Contribute to Risk of Type 2 Diabetes Mellitus: Evidence from a Meta-Analysis. J. Diabetes Res. 2016, 2016, 5412084. [Google Scholar] [CrossRef] [PubMed]
- Olza, J.; Ruperez, A.I.; Gil-Campos, M.; Leis, R.; Fernandez-Orth, D.; Tojo, R.; Cañete, R.; Gil, A.; Aguilera, C.M. Influence of FTO variants on obesity, inflammation and cardiovascular disease risk biomarkers in Spanish children: A case-control multicentre study. BMC Med. Genet. 2013, 14, 123. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Taylor, J.A. SNPinfo: Integrating GWAS and candidate gene information into functional SNP selection for genetic association studies. Nucleic Acids Res. 2009, 37, W600–W605. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Zhang, F.; Li, T.; Lu, M.; Wang, L.; Yue, W.; Zhang, D. MirSNP, a database of polymorphisms altering miRNA target sites, identifies miRNA-related SNPs in GWAS SNPs and eQTLs. BMC Genom. 2012, 13, 661. [Google Scholar] [CrossRef] [PubMed]
- RegSNP. Predicting Allele Specific Differences in Transcription Factor-DNA Binding. Available online: http://viis.abdn.ac.uk/regsnp/Home.aspx (accessed on 23 June 2016).
- Varela, L.; Horvath, T.L. Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012, 13, 1079–1086. [Google Scholar] [CrossRef] [PubMed]
- Berglund, E.D.; Vianna, C.R.; Donato, J., Jr.; Kim, M.H.; Chuang, J.C.; Lee, C.E.; Lauzon, D.A.; Lin, P.; Brule, L.J.; Scott, M.M.; et al. Direct leptin action on POMC neurons regulates glucose homeostasis and hepatic insulin sensitivity in mice. J. Clin. Investig. 2012, 122, 1000–1009. [Google Scholar] [CrossRef] [PubMed]
- Huo, L.; Gamber, K.; Greeley, S.; Silva, J.; Huntoon, N.; Leng, X.H.; Bjørbaek, C. Leptin-dependent control of glucose balance and locomotor activity by POMC neurons. Cell Metab. 2009, 9, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Paz-Fhilo, G.; Mastronardi, C.; Franco, C. Leptin: Molecular mechanisms, systemic pro-inflammatory effects, and clinical implications. Arq. Bras. Endocrinol. Metabol. 2012, 56, 597–607. [Google Scholar] [CrossRef]
- Hollensted, M.; Ahluwalia, T.S.; Have, C.T.; Grarup, N.; Fonvig, C.E.; Nielsen, T.R.; Trier, C.; Paternoster, L.; Pedersen, O.; Holm, J.C.; et al. Common variants in LEPR, IL6, AMD1, and NAMPT do not associate with risk of juvenile and childhood obesity in Danes: A case-control study. BMC Med. Genet. 2015, 16, 105. [Google Scholar] [CrossRef] [PubMed]
- Cole, T.J.; Bellizzi, M.C.; Flegal, K.M.; Dietz, W.H. Establishing a standard definition for child overweight and obesity worldwide: International survey. BMJ 2000, 320, 1240–1243. [Google Scholar] [CrossRef] [PubMed]
- Sobradillo, B.; Aguirre, A.; Aresti, U.; Bilbao, A.; Fernández-Ramos, C.; Lizárraga, A.; Lorenzo, H.; Madariaga, L.; Rica, I.; Ruiz, I.; et al. Curvas y Tablas de Crecimiento (Estudios Longitudinal y Transversal); Fundación Faustino Orbegozo Eizaguirre: Madrid, Spain, 2004. [Google Scholar]
- National High Blood Pressure Education Program Working Group on Hypertension Control in Children and Adolescents. Update on the 1987 Task Force Report on High Blood Pressure in Children and Adolescents: A Working Group Report from the National High Blood Pressure Education Program. Pediatrics 1996, 98, 649–658. [Google Scholar]
- Matthews, D.R.; Hosker, J.P.; Rudenski, A.S.; Naylor, B.A.; Treacher, D.F.; Turner, R.C. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [PubMed]
- Olza, J.; Gil-Campos, M.; Leis, R.; Rupérez, A.I.; Tojo, R.; Cañete, R.; Gil, A.; Aguilera, C.M. A gene variant of 11β-hydroxysteroid dehydrogenase type 1 is associated with obesity in children. Int. J. Obes. 2012, 36, 1558–1563. [Google Scholar] [CrossRef] [PubMed]
- Rupérez, A.I.; Olza, J.; Gil-Campos, M.; Leis, R.; Mesa, M.D.; Tojo, R.; Cañete, R.; Gil, A.; Aguilera, C.M. Are catalase -844A/G polymorphism and activity associated with childhood obesity? Antioxid. Redox Signal. 2013, 19, 1970–1975. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
| Normal-BMI | Obese | p | |
|---|---|---|---|
| n | 236 | 286 | |
| Anthropometry | |||
| Sex (M/F) | 133/103 | 146/140 | 0.252 |
| Age (y) | 9.72 ± 0.16 | 9.43 ± 0.15 | 0.188 |
| Tanner Stage (Prepuber/Puber) | |||
| Male | 113/20 | 122/24 | |
| Female | 83/20 | 108/32 | |
| Weight (kg) | 32.9 ± 0.7 | 55.9 ± 1.0 | <0.001 |
| Height (m) | 1.37 ± 0.01 | 1.41 ± 0.01 | 0.002 |
| BMI (kg/m2) | 17.14 ± 0.13 | 27.59 ± 0.24 | <0.001 |
| BMI z-Score | −0.17 ± 0.04 | 3.50 ± 0.08 | <0.001 |
| Waist circumference (cm) | 60.3 ± 0.5 | 84.0 ± 0.9 | <0.001 |
| Clinical and Metabolic Biomarkers | |||
| Systolic BP (mm Hg) | 98 ± 1 | 111 ± 1 | <0.001 |
| Diastolic BP (mm Hg) | 60 ± 1 | 69 ± 1 | <0.001 |
| Glucose (mg/dL) | 84 ± 1 | 85 ± 1 | 0.816 |
| Insulin (mU/L) | 5.89 ± 0.23 | 11.53 ± 0.52 | <0.001 |
| HOMA-IR | 1.26 ± 0.05 | 2.45 ± 0.12 | <0.001 |
| QUICKI | 0.383 ± 0.003 | 0.347 ± 0.002 | <0.001 |
| Triacylglycerols (mg/dL) | 55 ± 1 | 75 ± 2 | <0.001 |
| Apo-AI (mg/dL) | 149 ± 2 | 132 ± 2 | <0.001 |
| Apo-B (mg/dL) | 67 ± 1 | 71 ± 1 | 0.006 |
| Cholesterol (mg/dL) | 171 ± 2 | 165 ± 2 | 0.024 |
| HDL-c (mg/dL) | 64 ± 1 | 51 ± 1 | <0.001 |
| LDL-c (mg/dL) | 94 ± 2 | 97 ± 2 | 0.136 |
| AST (U/L) | 23.70 ± 0.48 | 21.23 ± 0.40 | <0.001 |
| ALT (U/L) | 16.80 ± 0.57 | 20.88 ± 0.51 | <0.001 |
| GGT (U/L) | 8.44 ± 0.27 | 10.90 ± 0.30 | <0.001 |
| Adiponectin (mg/L) | 28.23 ± 0.77 | 22.53 ± 0.66 | <0.001 |
| Resistin (μg/L) | 9.67 ± 0.34 | 11.77 ± 0.35 | <0.001 |
| Leptin (μg/L) | 4.30 ± 0.26 | 23.15 ± 0.87 | <0.001 |
| Inflammation Biomarkers | |||
| C-reactive protein (mg/L) | 0.97 ± 0.23 | 3.44 ± 0.25 | <0.001 |
| Interleukin 6 (ng/L) | 4.55 ± 0.54 | 7.03 ± 0.76 | 0.008 |
| Interleukin 8 (ng/L) | 1.57 ± 0.11 | 2.17 ± 0.15 | 0.002 |
| TNF-α (ng/L) | 3.04 ± 0.11 | 4.00 ± 0.13 | <0.001 |
| Cardiovascular Disease Risk Biomarkers | |||
| MMP-9 (μg/L) | 79.72 ± 3.17 | 87.98 ± 3.92 | 0.714 |
| MPO (μg/L) | 13.18 ± 1.18 | 21.70 ±1.73 | <0.001 |
| sE selectin (μg/L) | 22.91 ± 0.77 | 31.36 ± 1.06 | <0.001 |
| sICAM-1 (mg/L) | 0.153 ± 0.004 | 0.174 ± 0.005 | <0.001 |
| Active PAI-1 (μg/L) | 5.07 ± 0.26 | 11.92 ± 0.58 | <0.001 |
| Total PAI-1 (μg/L) | 18.82 ± 0.85 | 27.11 ± 1.12 | <0.001 |
| Polymorphism | Function | Allele 1/Allele 2 | Case | Control | Minor Allele | Minor Allele | OR (95% CI) | p | p a | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | 11 | 12 | 22 | Case | Control | |||||||
| rs11208659 | Intron | T/C | 240 | 44 | 2 | 173 | 61 | 2 | C | 0.084 | 0.138 | 0.54 (0.35–0.81) | 0.003 | 0.076 |
| rs11804091 | Intron | A/G | 193 | 76 | 8 | 181 | 46 | 2 | G | 0.166 | 0.109 | 1.64 (1.13–2.39) | 0.010 | 0.251 |
| rs10157275 | Intron | C/T | 195 | 83 | 8 | 184 | 46 | 6 | T | 0.173 | 0.123 | 1.53 (1.08–2.18) | 0.017 | 0.444 |
| rs9436303 | Intron | A/G | 162 | 100 | 24 | 152 | 73 | 11 | G | 0.259 | 0.201 | 1.36 (1.02–1.81) | 0.036 | 0.926 |
| rs1627238 | Intron | C/T | 186 | 87 | 13 | 167 | 60 | 5 | T | 0.198 | 0.151 | 1.40 (1.01–1.94) | 0.046 | 1 |
| rs17412175 | Intron | T/A | 82 | 146 | 58 | 57 | 120 | 58 | A | 0.458 | 0.502 | 0.82 (0.64–1.06) | 0.133 | 1 |
| rs9436739 | Intron | T/A | 231 | 53 | 2 | 178 | 56 | 2 | A | 0.100 | 0.127 | 0.74 (0.50–1.10) | 0.135 | 1 |
| rs1137101 | Gln223Arg | A/G | 85 | 135 | 65 | 76 | 117 | 41 | G | 0.465 | 0.425 | 1.16 (0.91–1.48) | 0.243 | 1 |
| rs6673591 | Intron | A/G | 83 | 129 | 74 | 55 | 121 | 60 | G | 0.484 | 0.511 | 0.89 (0.70–1.13) | 0.325 | 1 |
| rs17412723 | Intron | A/G | 71 | 155 | 60 | 58 | 114 | 62 | G | 0.481 | 0.509 | 0.88 (0.68–1.14) | 0.329 | 1 |
| rs6697315 | Intron | T/C | 126 | 125 | 35 | 92 | 113 | 30 | C | 0.341 | 0.368 | 0.88 (0.68–1.14) | 0.329 | 1 |
| rs6704167 | Intron | A/T | 92 | 138 | 56 | 68 | 114 | 52 | T | 0.437 | 0.466 | 0.88 (0.69–1.13) | 0.330 | 1 |
| rs8179183 | Lys656Asn | G/C | 195 | 82 | 9 | 153 | 72 | 10 | C | 0.175 | 0.196 | 0.87 (0.63–1.19) | 0.379 | 1 |
| rs1327118 | PRO | G/C | 73 | 143 | 59 | 62 | 121 | 42 | C | 0.475 | 0.456 | 1.10 (0.85–1.43) | 0.472 | 1 |
| rs1137100 | Lys109Arg | A/G | 155 | 112 | 19 | 133 | 89 | 14 | G | 0.262 | 0.248 | 1.09 (0.82–1.45) | 0.552 | 1 |
| rs3806318 | PRO | A/G | 158 | 104 | 24 | 116 | 101 | 16 | G | 0.266 | 0.285 | 0.92 (0.70–1.21) | 0.562 | 1 |
| rs970468 | Intron | T/G | 123 | 135 | 28 | 96 | 113 | 26 | G | 0.334 | 0.351 | 0.93 (0.71–1.22) | 0.588 | 1 |
| rs3790429 | Intron | A/T | 187 | 93 | 5 | 161 | 64 | 8 | T | 0.181 | 0.172 | 1.09 (0.78–1.51) | 0.630 | 1 |
| rs9436740 | Intron | A/T | 143 | 115 | 24 | 119 | 89 | 26 | T | 0.289 | 0.301 | 0.95 (0.73–1.24) | 0.704 | 1 |
| rs1475397 | Intron | C/T | 149 | 118 | 19 | 122 | 95 | 19 | T | 0.273 | 0.282 | 0.95 (0.72–1.25) | 0.712 | 1 |
| rs11585329 | Intron | T/G | 215 | 64 | 7 | 171 | 61 | 4 | T | 0.136 | 0.146 | 0.94 (0.66–1.33) | 0.718 | 1 |
| rs4655802 | Intron | A/G | 92 | 133 | 55 | 74 | 111 | 41 | G | 0.434 | 0.427 | 1.03 (0.80–1.32) | 0.828 | 1 |
| rs6678033 | Intron | G/A | 107 | 136 | 43 | 91 | 111 | 34 | A | 0.388 | 0.379 | 1.03 (0.80–1.33) | 0.829 | 1 |
| rs6672331 | Intron | G/C | 273 | 13 | 0 | 224 | 12 | 0 | C | 0.023 | 0.025 | 0.94 (0.42–2.12) | 0.886 | 1 |
| rs1137099 | Thr85Ala | A/ | 286 | 0 | 0 | 236 | 0 | 0 | 0 | 0 | - | - | - | |
| rs13306526 | Ile503Val | A/ | 286 | 0 | 0 | 236 | 0 | 0 | 0 | 0 | - | - | - | |
| Polymorphism | Allele 1/Allele 2 | Case | Control | Minor Allele | Minor Allele | OR (95% CI) | p | p a | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | 11 | 12 | 22 | Case | Control | ||||||
| Females | |||||||||||||
| rs11208659 | T/C | 113 | 26 | 1 | 72 | 3 | 1 | C | 0.100 | 0.155 | 0.56 (0.31–1.00) | 0.050 | 1 |
| rs11804091 | A/G | 87 | 42 | 4 | 82 | 19 | 0 | G | 0.188 | 0.094 | 2.73 (1.47–5.08) | 0.001 | 0.038 |
| rs10157275 | C/T | 99 | 37 | 4 | 84 | 17 | 2 | T | 0.161 | 0.102 | 1.77 (1.01–3.12) | 0.045 | 1 |
| rs9436303 | A/G | 77 | 52 | 11 | 68 | 30 | 5 | G | 0.264 | 0.194 | 1.49 (0.95–2.32) | 0.080 | 1 |
| rs1627238 | C/T | 93 | 41 | 6 | 75 | 23 | 2 | T | 0.189 | 0.135 | 1.59 (0.95–2.32) | 0.079 | 1 |
| rs1327118 | G/C | 32 | 73 | 30 | 29 | 54 | 12 | C | 0.493 | 0.411 | 1.53 (1.01–2.32) | 0.048 | 1 |
| Males | |||||||||||||
| rs11208659 | T/C | 127 | 18 | 1 | 101 | 31 | 1 | C | 0.068 | 0.124 | 0.50 (0.27–0.92) | 0.026 | 0.681 |
| rs11804091 | A/G | 106 | 34 | 4 | 99 | 27 | 2 | G | 0.146 | 0.121 | 1.23 (0.76–2.02) | 0.837 | 1 |
| rs10157275 | C/T | 96 | 46 | 4 | 100 | 29 | 4 | T | 0.185 | 0.139 | 1.40 (0.89–2.21) | 0.147 | 1 |
| rs9436303 | A/G | 85 | 48 | 13 | 84 | 43 | 6 | G | 0.253 | 0.207 | 1.28 (0.87–1.87) | 0.218 | 1 |
| rs1627238 | C/T | 93 | 46 | 7 | 92 | 37 | 3 | T | 0.206 | 0.163 | 1.33 (0.86–2.04) | 0.202 | 1 |
| rs1327118 | G/C | 41 | 70 | 29 | 33 | 67 | 30 | C | 0.457 | 0.489 | 0.88(0.63–1.24) | 0.465 | 1 |
| Biomarkers | AA | AG | GG | β (95% CI) | p | p a |
|---|---|---|---|---|---|---|
| n | 169 | 61 | 4 | |||
| Anthropometry | ||||||
| Height (m) | 1.37 ± 0.01 | 1.41 ± 0.02 | 1.35 ± 0.05 | −0.014 (−0.025, −0.004) | 0.010 | -- |
| Weight (kg) | 43.1 ± 1.4 | 49.9 ± 2.5 | 50.5 ± 7.9 | 6.1 (2.6, 9.6) | 0.001 | -- |
| BMI (kg/m2) | 22.44 ± 0.47 | 24.41 ± 0.78 | 27.33 ± 2.15 | 2.20 (0.70, 3.70) | 0.004 | -- |
| BMI z-Score | 1.57 ± 0.15 | 2.08 ± 0.23 | 3.51 ± 0.45 | 0.70 (0.22, 1.18) | 0.004 | -- |
| Waist circumference (cm) | 71.56 ± 1.31 | 76.07 ± 2.14 | 80.75 ± 2.39 | 5.46 (2.62, 9.59) | 0.055 | 0.667 |
| Clinical and Metabolic Biomarkers | ||||||
| Systolic BP (mm Hg) | 104 ± 1 | 108 ± 2 | 120 ± 3 | 5.56 (1.97, 9.14) | 0.003 | 0.092 |
| Diastolic BP (mm Hg) | 65 ± 1 | 66 ± 1 | 73 ± 6 | 2.49 (−0.51, 5.49) | 0.105 | 0.569 |
| Glucose (mg/dL) | 84 ± 1 | 85 ± 1 | 79 ± 2 | −0.08 (−1.94, 1.78) | 0.934 | 0.989 |
| Insulin (mU/L) | 9.11 ± 0.60 | 12.64 ± 1.15 | 10.95 ± 3.25 | 0.14 (0.07, 0.21) | 0.0001 | 0.004 |
| HOMA-IR | 1.91 ± 0.13 | 2.70 ± 0.28 | 2.17 ± 0.69 | 0.14 (0.07, 0.22) | 0.0002 | 0.006 |
| QUICKI | 0.367 ± 0.003 | 0.342 ± 0.004 | 0.349 ± 0.014 | −0.019 (−0.028, −0.009) | 0.0001 | 0.005 |
| Triacylglycerols (mg/dL) | 69 ± 3 | 70 ± 4 | 130 ± 32 | 8.50 (−0.17, 17.17) | 0.056 | 0.376 |
| Apo-AI (mg/dL) | 139 ± 2 | 131 ± 3 | 122 ± 9 | −8.19 (−15.01, −1.37) | 0.019 | 0.129 |
| Cholesterol (mg/dL) | 168 ± 2 | 169 ± 4 | 180 ± 14 | 2.01 (−5.11, 0.55) | 0.604 | 0.334 |
| HDL-c (mg/dL) | 55 ± 2 | 52 ± 2 | 62 ± 17 | −1.83 (−5.68, 2.01) | 0.350 | 0.649 |
| Adiponectin (mg/L) | 26.67 ± 0.95 | 20.96 ± 1.48 | 23.33 ± 2.30 | −4.77 (−7.94, −1.60) | 0.004 | 0.046 |
| Leptin (μg/L) | 13.66 ± 1.03 | 19.01 ± 2.13 | 19.81 ± 3.69 | 5.10 (0.04, 1.06) | 0.006 | 0.314 |
| Inflammation Biomarkers | ||||||
| C-reactive protein (mg/L) | 2.29 ± 0.41 | 3.42 ± 0.52 | 1.65 ± 0.61 | 0.84 (−0.45, 2.13) | 0.202 | 0.633 |
| IL-6 (ng/L) | 6.03 ± 0.89 | 5.38 ± 0.99 | 18.16 ± 9.07 | 0.92 (−1.91, 3.77) | 0.522 | 0.737 |
| IL-8 (ng/L) | 1.80 ± 0.13 | 1.79 ± 0.25 | 3.63± 1.51 | 0.21 (−0.26, 0.69) | 0.381 | 0.637 |
| TNF-α (ng/L) | 3.26 ± 0.15 | 3.84 ± 0.31 | 4.02 ± 0.94 | 0.55 (0.04, 1.06) | 0.035 | 0.090 |
| Cardiovascular Disease Risk Biomarkers | ||||||
| MMP-9 (µg/L) | 84.18 ± 4.44 | 78.82 ± 6.59 | 75.67 ± 13.56 | −5.36 (−19.92, 9.11) | 0.494 | 0.489 |
| MPO (µg/L) | 17.27 ± 1.47 | 21.59 ± 4.28 | 22.05 ± 7.14 | 3.47 (−2.63, 9.58) | 0.266 | 0.572 |
| sE-Selectin (µg/L) | 26.78 ± 1.22 | 29.67 ± 2.44 | 21.99 ± 5.74 | 2.54 (−1.89, 6.97) | 0.263 | 0.518 |
| sICAM-1 (mg/L) | 0.164 ± 0.005 | 0.160 ± 0.009 | 0.218 ± 0.047 | 0.002 (−0.016, 0.021) | 0.766 | 0.889 |
| Active PAI-1 (µg/L) | 9.51 ± 0.69 | 9.63 ± 1.12 | 15.55 ± 6.81 | 0.89 (−1.47, 3.26) | 0.461 | 0.441 |
| Total PAI-1 (µg/L) | 23.75 ± 1.29 | 23.56 ± 2.54 | 28.67 ± 9.17 | 0.62 (−3.98, 5.23) | 0.713 | 0.707 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olza, J.; Rupérez, A.I.; Gil-Campos, M.; Leis, R.; Cañete, R.; Tojo, R.; Gil, Á.; Aguilera, C.M. Leptin Receptor Gene Variant rs11804091 Is Associated with BMI and Insulin Resistance in Spanish Female Obese Children: A Case-Control Study. Int. J. Mol. Sci. 2017, 18, 1690. https://doi.org/10.3390/ijms18081690
Olza J, Rupérez AI, Gil-Campos M, Leis R, Cañete R, Tojo R, Gil Á, Aguilera CM. Leptin Receptor Gene Variant rs11804091 Is Associated with BMI and Insulin Resistance in Spanish Female Obese Children: A Case-Control Study. International Journal of Molecular Sciences. 2017; 18(8):1690. https://doi.org/10.3390/ijms18081690
Chicago/Turabian StyleOlza, Josune, Azahara I. Rupérez, Mercedes Gil-Campos, Rosaura Leis, Ramón Cañete, Rafael Tojo, Ángel Gil, and Concepción M. Aguilera. 2017. "Leptin Receptor Gene Variant rs11804091 Is Associated with BMI and Insulin Resistance in Spanish Female Obese Children: A Case-Control Study" International Journal of Molecular Sciences 18, no. 8: 1690. https://doi.org/10.3390/ijms18081690
APA StyleOlza, J., Rupérez, A. I., Gil-Campos, M., Leis, R., Cañete, R., Tojo, R., Gil, Á., & Aguilera, C. M. (2017). Leptin Receptor Gene Variant rs11804091 Is Associated with BMI and Insulin Resistance in Spanish Female Obese Children: A Case-Control Study. International Journal of Molecular Sciences, 18(8), 1690. https://doi.org/10.3390/ijms18081690