Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species
Abstract
:1. Introduction
2. Results and Discussion
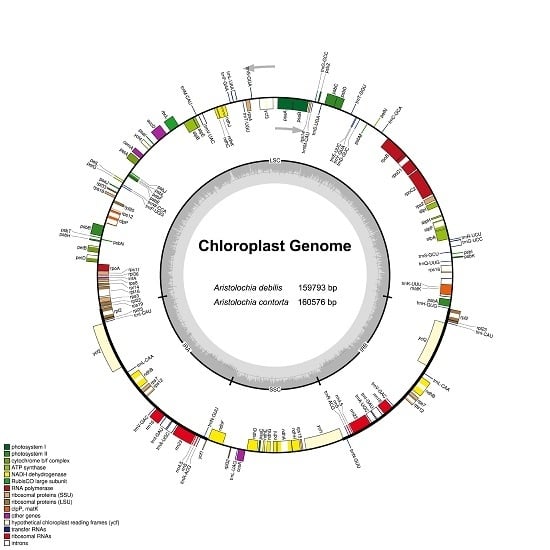
2.1. The Chloroplast Genome Structures of A. debilis and A. contorta
2.2. IR Contraction and Expansion
2.3. Codon Usage and RNA Editing Sites
2.4. Repeat Structure and Simple Sequence Repeats Analyses
2.5. Comparative Genomic Analysis
2.6. Phylogenetic Analyses
3. Materials and Methods
3.1. Plant Material, DNA Extraction, and Sequencing
3.2. Chloroplast Genome Assembly and Annotation
3.3. Genome Structure Analyses and Genome Comparison
3.4. Phylogenetic Analyses
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| LSC | Large single copy |
| SSC | Small single copy |
| IR | Inverted repeat |
| MP | Maximum parsimony |
| SSR | Simple sequence repeats |
| ATP | Adenosine triphosphate |
| NADH | Nicotinamide adenine dinucleotide |
References
- Chinese Pharmacopoeia Commission. The Chinese Pharmacopoeia; Chemical Industry Press: Beijing, China, 2015; pp. 51–52. [Google Scholar]
- Chen, C.X. Studies on the chemical constituents from the fruit of Aristolochia debilis. J. Chin. Med. Mater. 2010, 33, 1260–1261. [Google Scholar]
- Xu, Y.; Shang, M.; Ge, Y.; Wang, X.; Cai, S. Chemical constituent from fruit of Aristolochia contorta. Chin. J. Chin. Mater. Medica 2010, 35, 2862. [Google Scholar]
- Mix, D.B.; Guinaudeau, H.; Shamma, M. The aristolochic acids and aristolactams. J. Nat. Prod. 1982, 45, 657–666. [Google Scholar] [CrossRef]
- Arlt, V.M.; Stiborova, M.; Schmeiser, H.H. Aristolochic acid as a probable human cancer hazard in herbal remedies: A review. Mutagenesis 2002, 17, 265. [Google Scholar] [CrossRef] [PubMed]
- Schmeiser, H.H.; Janssen, J.W.; Lyons, J.; Scherf, H.R.; Pfau, W.; Buchmann, A.; Bartram, C.R.; Wiessler, M. Aristolochic acid activates RAS genes in rat tumors at deoxyadenosine residues. Cancer Res. 1990, 50, 5464–5469. [Google Scholar] [PubMed]
- Chen, L.; Mei, N.; Yao, L.; Chen, T. Mutations induced by carcinogenic doses of aristolochic acid in kidney of big blue transgenic rats. Toxicol. Lett. 2006, 165, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.L.; Chen, K.J.; Shih, P.H.; Lu, L.Y.; Hung, C.F.; Lin, W.C.; Yesong, G.J. Chronic renal failure rats are highly sensitive to aristolochic acids, which are nephrotoxic and carcinogenic agents. Cancer Lett. 2006, 232, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Cosyns, J.P.; Goebbels, R.M.; Liberton, V.; Schmeiser, H.H.; Bieler, C.A.; Bernard, A.M. Chinese herbs nephropathy-associated slimming regimen induces tumours in the forestomach but no interstitial nephropathy in rats. Arch. Toxicol. 1998, 72, 738–743. [Google Scholar] [CrossRef] [PubMed]
- Hoang, M.L.; Chen, C.H.; Sidorenko, V.S.; He, J.; Dickman, K.G.; Yun, B.H.; Moriya, M.; Niknafs, N.; Douville, C.; Karchin, R. Mutational signature of aristolochic acid exposure as revealed by whole-exome sequencing. Sci. Transl. Med. 2013, 5. [Google Scholar] [CrossRef] [PubMed]
- Balachandran, P.; Wei, F.; Lin, R.C.; Khan, I.A.; Pasco, D.S. Structure activity relationships of aristolochic acid analogues: Toxicity in cultured renal epithelial cells. Kidney Int. 2005, 67, 1797. [Google Scholar] [CrossRef] [PubMed]
- Tsai, D.M.; Kang, J.J.; Lee, S.S.; Wang, S.Y.; Tsai, I.; Chen, G.Y.; Liao, H.W.; Li, W.C.; Kuo, C.H.; Tseng, Y.J. Metabolomic analysis of complex Chinese remedies: Examples of induced nephrotoxicity in the mouse from a series of remedies containing aristolochic acid. Evid. Based Compl. Alt. 2013, 2013, 263757. [Google Scholar] [CrossRef] [PubMed]
- Grollman, A.P.; Shibutani, S.; Moriya, M.; Miller, F.; Wu, L.; Moll, U.; Suzuki, N.; Fernandes, A.; Rosenquist, T.; Medverec, Z.; et al. Aristolochic acid and the etiology of endemic (Balkan) nephropathy. Proc. Natl. Acad. Sci. USA 2007, 104, 12129–12134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, G.M.; Tagore, R.; Cook, T.; Gower, P.; Pusey, C.D. Nephropathy caused by Chinese herbs in the UK. Lancet 1999, 354, 481–482. [Google Scholar] [CrossRef]
- Vanherweghem, J.L.; Tielemans, C.; Abramowicz, D.; Depierreux, M.; Vanhaelen-Fastre, R.; Vanhaelen, M.; Dratwa, M.; Richard, C.; Vandervelde, D.; Verbeelen, D.; et al. Rapidly progressive interstitial renal fibrosis in young women: Association with slimming regimen including Chinese herbs. Lancet 1993, 341, 387–391. [Google Scholar] [CrossRef]
- Dong, W.; Xu, C.; Cheng, T.; Lin, K.; Zhou, S. Sequencing angiosperm plastid genomes made easy: A complete set of universal primers and a case study on the phylogeny of Saxifragales. Genome Biol. Evol. 2013, 5, 989–997. [Google Scholar] [CrossRef] [PubMed]
- Leliaert, F.; Smith, D.R.; Moreau, H.; Herron, M.D.; Verbruggen, H.; Delwiche, C.F.; Clerck, O.D. Phylogeny and molecular evolution of the green algae. Crit. Rev. Plant Sci. 2012, 31, 1–46. [Google Scholar] [CrossRef]
- Raman, G.; Park, S. Analysis of the complete chloroplast genome of a medicinal plant, Dianthus superbus var. longicalyncinus, from a comparative genomics perspective. PLoS ONE 2015, 10. [Google Scholar] [CrossRef]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; Depamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [PubMed]
- Wolfe, K.H.; Li, W.H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast and nuclear DNA. Proc. Natl. Acad. Sci. USA 1988, 84, 9054–9058. [Google Scholar] [CrossRef]
- Smith, D.R.; Keeling, P.J. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc. Natl. Acad. Sci. USA 2015, 112, 10177–10184. [Google Scholar] [CrossRef] [PubMed]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1,000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Wicke, S.; Schneeweiss, G.M.; Müller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant. Mol. Boil. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.Y.; Wu, H.; Wang, Y.L.; Gao, L.M.; Zhang, S.Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhan, D.F.; Jia, X.; Mei, W.L.; Dai, H.F.; Chen, X.T.; Peng, S.Q. Complete chloroplast genome sequence of Aquilaria sinensis (lour.) gilg and evolution analysis within the Malvales order. Front. Plant Sci. 2016, 7, 280. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.H.; Chan, M.T.; Liao, D.C.; Chentran, H.; Yiwei, L.; Daniell, H.; Duvall, M.R.; Lin, C.S. Complete chloroplast genome of Oncidium Gower Ramsey and evaluation of molecular markers for identification and breeding in Oncidiinae. BMC Plant Biol. 2010, 10, 68. [Google Scholar] [CrossRef] [PubMed]
- Bedbrook, J.R.; Bogorad, L. Endonuclease recognition sites mapped on Zea mays chloroplast DNA. Proc. Natl. Acad. Sci. USA 1976, 73, 4309–4313. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Ohme, M.; Tanaka, M.; Wakasugi, T.; Hayshida, N.; Matsubayasha, T.; Zaita, N.; Chunwongse, J.; Obokata, J.; Yamaguchi-Shinozaki, K. The complete nucleotide sequence of the tobacco chloroplast genome. EMBO J. 1986, 4, 111–148. [Google Scholar] [CrossRef]
- NCBI, Genome. Available online: https://www.ncbi.nlm.nih.gov/genome/browse/?report=5 (accessed on 30 June 2017).
- The Editorial Committee of Flora of China. Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2003; Volume 5, pp. 246–269. [Google Scholar]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H.; Sun, C.; Li, X.; Li, C. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Clegg, M.T.; Gaut, B.S.; Learn, G.H., Jr.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Zhang, X.; Liu, G.; Yin, Y.; Chen, K.; Yun, Q.; Zhao, D.; Al-Mssallem, I.S.; Yu, J. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS ONE 2010, 5. [Google Scholar] [CrossRef] [PubMed]
- Yi, D.K.; Kim, K.J. Complete chloroplast genome sequences of important oilseed crop Sesamum indicum L. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Feng, D.; Song, G.; Wei, X.; Chen, L.; Wu, X.; Li, X.; Zhu, Z. The first intron of rice EPSP synthase enhances expression of foreign gene. Sci. China Life Sci. 2003, 46, 561–569. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.J.; Lee, H.L. Complete chloroplast genome sequences from Korean ginseng (Panax schinseng nees) and comparative analysis of sequence evolution among 17 vascular plants. DNA Res. 2004, 11, 247. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, T.C.; Qiao, Q.; Ren, Z.; Zhao, J.; Yonezawa, T.; Hasegawa, M.; Crabbe, M.J.; Li, J.; Zhong, Y. Complete chloroplast genome sequence of holoparasite Cistanche deserticola (Orobanchaceae) reveals gene loss and horizontal gene transfer from its host Haloxylon ammodendron (Chenopodiaceae). PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raubeson, L.A.; Peery, R.; Chumley, T.W.; Dziubek, C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Comparative chloroplast genomics: Analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genom. 2007, 8, 174. [Google Scholar] [CrossRef] [PubMed]
- Huotari, T.; Korpelainen, H. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene 2012, 508, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Zuo, L.H.; Shang, A.Q.; Zhang, S.; Yu, X.Y.; Ren, Y.C.; Yang, M.S.; Wang, J.M. The first complete chloroplast genome sequences of Ulmus species by de novo sequencing: Genome comparative and taxonomic position analysis. PLoS ONE 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Powell, W.; Morgante, M.; McDevitt, R.; Vendramin, G.G.; Rafalski, J.A. Polymorphic simple sequence repeat regions in chloroplast genomes: Applications to the population genetics of pines. Proc. Natl. Acad. Sci. USA 1995, 92, 7759–7763. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Jia, H.; Li, X.; Chai, M.; Jia, H.; Chen, Z.; Wang, G.; Chai, C.; Weg, E.V.D.; Gao, Z. Development of simple sequence repeat (SSR) markers from a genome survey of Chinese bayberry (Myrica rubra). BMC Genomics 2012, 13, 201. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Wang, S.; Zhou, S.L. Polymorphic chloroplast microsatellite loci in Nelumbo (Nelumbonaceae). Am. J. Bot. 2012, 99, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.H.; Zhang, J.J.; Yao, X.H.; Huang, H.W. Chloroplast microsatellite markers in Liriodendron tulipifera (Magnoliaceae) and cross-species amplification in L. chinense. Am. J. Bot. 2011, 98, 123–126. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Müller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.J.; Bell, C.D.; Soltis, P.S.; Soltis, D.E. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl. Acad. Sci. USA 2007, 104, 19363–19368. [Google Scholar] [CrossRef] [PubMed]
- Murata, J.; Ohi, T.; Wu, S.; Darnaedi, D.; Sugawara, T.; Nakanishi, T.; Murata, H. Molecular phylogeny of Aristolochia (Aristolochiaceae) inferred from matK sequences. Apg Acta Phyto. Geo. 2001, 52, 75–83. [Google Scholar]
- Ohi-Toma, T.; Sugawara, T.; Murata, H.; Wanke, S.; Neinhuis, C.; Jin, M. Molecular phylogeny of Aristolochia sensu lato (Aristolochiaceae) based on sequences of rbcL, matK, and phyA genes, with special reference to differentiation of chromosome numbers. Syst. Bot. 2006, 31, 481–492. [Google Scholar] [CrossRef]
- Silva-Brandão, K.L.; Solferini, V.N.; Trigo, J.R. Chemical and phylogenetic relationships among Aristolochia L. (Aristolochiaceae) from southeastern Brazil. Biochem. Syst. Ecol. 2006, 34, 291–302. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Luo, R.; Liu, B.; Xie, Y.; Li, Z.; Huang, W.; Yuan, J.; He, G.; Chen, Y.; Pan, Q.; Liu, Y.; et al. SOAPdenovo2: An empirically improved memory-efficient short-read de novo assembler. GigaScience 2012, 1, 18. [Google Scholar] [CrossRef] [PubMed]
- Wyman, S.K.; Jansen, R.K.; Boore, J.L. Automatic annotation of organellar genomes with DOGMA. Bioinformatics 2004, 20, 3252–3255. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Bock, R. Organellargenomedraw (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Sharp, P.M.; Li, W.H. The codon Adaptation Index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987, 15, 1281–1295. [Google Scholar] [CrossRef] [PubMed]
- Mower, J.P. The PREP suite: Predictive RNA editors for plant mitochondrial genes, chloroplast genes and user-defined alignments. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Comput. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. Reputer: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Misa-Microsatellite Identification Tool. Available online: http://pgrc.ipk-gatersleben.de/misa/ (accessed on 2 June 2017).
- Li, X.W.; Gao, H.H.; Wang, Y.T.; Song, J.Y.; Henry, R.; Wu, H.Z.; Hu, Z.G.; Hui, Y.; Luo, H.M.; Luo, K. Complete chloroplast genome sequence of Magnolia grandiflora and comparative analysis with related species. Sci. China Life Sci. 2013, 56, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Swofford, D.L. PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods); Version 4.0b10; Sinauer Associates: Sunderland, MA, USA, 2002. [Google Scholar]







| Species | Regions | Positions | T(U) (%) | C (%) | A (%) | G (%) | Length (bp) |
|---|---|---|---|---|---|---|---|
| A. debilis | LSC | - | 32.2 | 18.7 | 31.2 | 17.9 | 89,609 |
| SSC | - | 34.0 | 17.4 | 33.2 | 15.5 | 19,834 | |
| IRa | - | 28.4 | 22.4 | 28.3 | 21.0 | 25,175 | |
| IRb | - | 28.3 | 21.0 | 28.4 | 22.4 | 25,175 | |
| Total | - | 31.2 | 19.5 | 30.5 | 18.8 | 159,793 | |
| CDS | - | 30.9 | 18.1 | 30.2 | 20.8 | 78,717 | |
| - | 1st position | 23.5 | 18.8 | 30.5 | 27.2 | 26,239 | |
| - | 2nd position | 32.2 | 20.5 | 29.2 | 18.1 | 26,239 | |
| - | 3rd position | 36.9 | 15.1 | 31.1 | 17.0 | 26,239 | |
| A. contorta | LSC | - | 32.2 | 18.7 | 31.2 | 17.8 | 89,781 |
| SSC | - | 33.9 | 17.4 | 33.3 | 15.4 | 19,877 | |
| IRa | - | 28.4 | 22.4 | 28.2 | 21.0 | 25,459 | |
| IRb | - | 28.2 | 21.0 | 28.4 | 22.4 | 25,459 | |
| Total | - | 31.2 | 19.5 | 30.6 | 18.8 | 160,576 | |
| CDS | - | 30.9 | 18.1 | 30.3 | 20.7 | 78,765 | |
| - | 1st position | 23.5 | 18.8 | 30.5 | 27.2 | 26,255 | |
| - | 2nd position | 32.2 | 20.6 | 29.2 | 18.1 | 26,255 | |
| - | 3rd position | 37.0 | 15.0 | 31.1 | 16.9 | 26,255 |
| No. | Group of Genes | Gene names | Amount |
|---|---|---|---|
| 1 | Photosystem I | psaA, psaB, psaC, psaI, psaJ | 5 |
| 2 | Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | 15 |
| 3 | Cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | 6 |
| 4 | ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | 6 |
| 5 | NADH dehydrogenase | ndhA *, ndhB *(×2)1, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12(1) |
| 6 | RubisCO large subunit | rbcL | 1 |
| 7 | RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | 4 |
| 8 | Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7(×2), rps8, rps11, rps12 **(×2), rps14, rps15, rps16 *, rps18, rps19 | 14(2) |
| 9 | Ribosomal proteins (LSU) | rpl2 *(×2), rpl14, rpl16 *, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 | 11(2) |
| 10 | Proteins of unknown function | ycf1, ycf2(×2), ycf3 **, ycf4 | 5(1) |
| 11 | Transfer RNAs | 37 tRNAs (6 contain an intron, 7 in the IRs) | 37(7) |
| 12 | Ribosomal RNAs | rrn4.5(×2), rrn5(×2), rrn16(×2), rrn23(×2) | 8(4) |
| 13 | Other genes | accD, clpP **, matK, ccsA, cemA, infA | 6 |
| Species | Gene | Location | Exon I (bp) | Intron I (bp) | Exon II (bp) | Intron II (bp) | Exon III (bp) |
|---|---|---|---|---|---|---|---|
| A. debilis | atpF | LSC | 145 | 805 | 410 | - | - |
| clpP | LSC | 71 | 781 | 292 | 678 | 255 | |
| ndhA | SSC | 552 | 1090 | 540 | - | - | |
| ndhB | IR | 777 | 705 | 756 | - | - | |
| petB | LSC | 6 | 214 | 642 | - | - | |
| petD | LSC | 6 | 485 | 476 | - | - | |
| rpl16 | LSC | 8 | 1065 | 403 | - | - | |
| rpl2 | IR | 391 | 657 | 431 | - | - | |
| rpoC1 | LSC | 430 | 776 | 1622 | - | - | |
| rps12 | LSC | 114 | - | 232 | 536 | 26 | |
| rps16 | LSC | 46 | 853 | 191 | - | - | |
| trnA-UGC | IR | 38 | 809 | 35 | - | - | |
| trnG-UCC | LSC | 24 | 761 | 48 | - | - | |
| trnI-GAU | IR | 37 | 937 | 35 | - | - | |
| trnK-UUU | LSC | 37 | 2658 | 35 | - | - | |
| trnL-UAA | LSC | 35 | 521 | 50 | - | - | |
| trnV-UAC | LSC | 39 | 597 | 37 | - | - | |
| ycf3 | LSC | 126 | 777 | 228 | 753 | 147 | |
| A. contorta | atpF | LSC | 145 | 771 | 410 | - | - |
| clpP | LSC | 71 | 821 | 292 | 664 | 255 | |
| ndhA | SSC | 552 | 1091 | 540 | - | - | |
| ndhB | IR | 777 | 716 | 756 | - | - | |
| petB | LSC | 6 | 214 | 642 | - | - | |
| petD | LSC | 7 | 485 | 476 | - | - | |
| rpl16 | LSC | 8 | 1088 | 403 | - | - | |
| rpl2 | IR | 391 | 657 | 431 | - | - | |
| rpoC1 | LSC | 430 | 776 | 1619 | - | - | |
| rps12 | LSC | 114 | - | 232 | 536 | 26 | |
| rps16 | LSC | 46 | 832 | 221 | - | - | |
| trnA-UGC | IR | 38 | 809 | 35 | - | - | |
| trnG-UCC | LSC | 24 | 751 | 48 | - | - | |
| trnI-GAU | IR | 37 | 938 | 35 | - | - | |
| trnK-UUU | LSC | 37 | 2648 | 35 | - | - | |
| trnL-UAA | LSC | 35 | 552 | 50 | - | - | |
| trnV-UAC | LSC | 39 | 605 | 37 | - | - | |
| ycf3 | LSC | 126 | 764 | 228 | 760 | 147 |
| SSR Type | Repeat Unit | Amount | Ratio (%) | ||
|---|---|---|---|---|---|
| A. debilis | A. contorta | A. debilis | A. contorta | ||
| Mono | A/T | 78 | 91 | 96.3 | 94.8 |
| C/G | 3 | 5 | 3.7 | 5.2 | |
| Di | AC/GT | 0 | 1 | 0 | 3.6 |
| AG/CT | 0 | 1 | 0 | 3.6 | |
| AT/TA | 19 | 26 | 100 | 92.8 | |
| Tri | AAC/GTT | 1 | 1 | 10 | 8.3 |
| AAG/CTT | 1 | 1 | 10 | 8.3 | |
| ATC/ATG | 1 | 0 | 10 | 0 | |
| AAT/ATT | 7 | 10 | 70 | 83.4 | |
| Tetra | AAAC/GTTT | 2 | 2 | 16.7 | 14.3 |
| AAAT/ATTT | 4 | 5 | 33.3 | 35.7 | |
| AATC/ATTG | 1 | 1 | 8.3 | 7.1 | |
| AGAT/ATCT | 2 | 1 | 16.7 | 7.1 | |
| AATT/AATT | 0 | 1 | 0 | 7.1 | |
| ACAT/ATGT | 0 | 1 | 0 | 7.1 | |
| AACT/AGTT | 1 | 1 | 8.3 | 7.1 | |
| AATG/ATTC | 2 | 2 | 16.7 | 14.3 | |
| Penta | AATAT/ATATT | 2 | 2 | 33.3 | 50 |
| AAATT/AATTT | 1 | 0 | 16.7 | 0 | |
| AAATC/ATTTG | 1 | 0 | 16.7 | 0 | |
| AACAT/ATGTT | 0 | 1 | 0 | 25 | |
| AAAAT/ATTTT | 2 | 1 | 33.3 | 25 | |
| Hexa | AAATAG/ATTTCT | 0 | 1 | 0 | 50 |
| ACATAT/ATATGT | 0 | 1 | 0 | 50 | |
| ACTGAT/AGTATC | 1 | 0 | 100 | 0 | |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, J.; Chen, X.; Cui, Y.; Sun, W.; Li, Y.; Wang, Y.; Song, J.; Yao, H. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. Int. J. Mol. Sci. 2017, 18, 1839. https://doi.org/10.3390/ijms18091839
Zhou J, Chen X, Cui Y, Sun W, Li Y, Wang Y, Song J, Yao H. Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. International Journal of Molecular Sciences. 2017; 18(9):1839. https://doi.org/10.3390/ijms18091839
Chicago/Turabian StyleZhou, Jianguo, Xinlian Chen, Yingxian Cui, Wei Sun, Yonghua Li, Yu Wang, Jingyuan Song, and Hui Yao. 2017. "Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species" International Journal of Molecular Sciences 18, no. 9: 1839. https://doi.org/10.3390/ijms18091839
APA StyleZhou, J., Chen, X., Cui, Y., Sun, W., Li, Y., Wang, Y., Song, J., & Yao, H. (2017). Molecular Structure and Phylogenetic Analyses of Complete Chloroplast Genomes of Two Aristolochia Medicinal Species. International Journal of Molecular Sciences, 18(9), 1839. https://doi.org/10.3390/ijms18091839