Visualization of Tau–Tubulin Interaction in a Living Cell Using Bifluorescence Complementation Technique
Abstract
:1. Introduction
2. Results and Discussion
2.1. Determination of BiFC-Labeling Site for Tubulin
2.2. Establishment of a Stable Tau-Tubulin BiFC Cell Line
2.3. Live-Cell Investigation of Tau–Tubulin Interaction upon the Treatment with Microtubule-Disrupting Agents
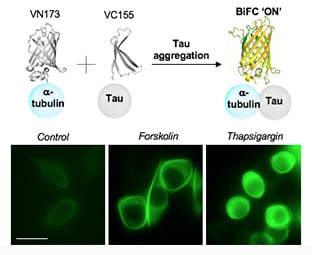
2.4. Increased Tau–Tubulin Interaction upon the Treatment of Tau Aggregation Inducters
2.5. Discussion
3. Experimental Section
3.1. Construction of Tubulin-BiFC Plasmids
3.2. Transient Transfection and BiFC-Image Acquisition
3.3. Establishment of Tau-Tubulin BiFC Stable Cell Line
3.4. Cell Culture and Compound Treatment
3.5. BiFC-Image Analysis
3.6. Immunofluorescence Stain
3.7. Immunoblot Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Barghorn, S.; Mandelkow, E. Toward a unified scheme for the aggregation of tau into Alzheimer paired helical filaments. Biochemistry 2002, 41, 14885–14896. [Google Scholar] [CrossRef] [PubMed]
- Brandt, R.; Hundelt, M.; Shahani, N. Tau alteration and neuronal degeneration in tauopathies: Mechanisms and models. Biochim. Biophys. Acta 2005, 1739, 331–354. [Google Scholar] [CrossRef] [PubMed]
- Berk, A.; Zipursky, S.; Lodish, H. Molecular Cell Biology, 4th ed.; W.H. Freeman: New York, NY, USA, 2000. [Google Scholar]
- Ballatore, C.; Lee, V.M.; Trojanowski, J.Q. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat. Rev. Neurosci. 2007, 8, 663–672. [Google Scholar] [CrossRef] [PubMed]
- Lee, V.M.; Goedert, M.; Trojanowski, J.Q. Neurodegenerative tauopathies. Annu. Rev. Neurosci. 2001, 24, 1121–1159. [Google Scholar] [CrossRef] [PubMed]
- Heald, R.; Nogales, E. Microtubule dynamics. J. Cell Sci. 2002, 115, 3–4. [Google Scholar] [PubMed]
- Kadavath, H.; Hofele, R.V.; Biernat, J.; Kumar, S.; Tepper, K.; Urlaub, H.; Mandelkow, E.; Zweckstetter, M. Tau stabilizes microtubules by binding at the interface between tubulin heterodimers. Proc. Natl. Acad. Sci. USA 2015. [Google Scholar] [CrossRef] [PubMed]
- Butner, K.; Kirschner, M.W. Tau protein binds to microtubules through a flexible array of distributed weak sites. J. Cell Biol. 1991, 115, 717–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elbaum-Garfinkle, S.; Cobb, G.; Compton, J.T.; Li, X.-H.; Rhoades, E. Tau mutants bind tubulin heterodimers with enhanced affinity. Proc. Natl. Acad. Sci. USA 2014, 111, 6311–6316. [Google Scholar] [CrossRef] [PubMed]
- Puig, B.; Ferrer, I.; Luduena, R.; Avila, J. βII-tubulin and phospho-tau aggregates in Alzheimer’s disease and Pick’s disease. J. Alzheimers Dis. 2005, 7, 213–220. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, K.; Liu, F.; Gong, C.-X. Tau and neurodegenerative disease: The story so far. Nat. Rev. Neurol. 2016, 12, 15. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Brady, S.T. Post-translational modifications of tubulin: Pathways to functional diversity of microtubules. Trends Cell Biol. 2015, 25, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-H.; Rhoades, E. Heterogeneous tau-tubulin complexes accelerate microtubule polymerization. Biophys. J. 2017, 112, 2567–2574. [Google Scholar] [CrossRef] [PubMed]
- Tak, H.; Haque, M.M.; Kim, M.J.; Lee, J.H.; Baik, J.H.; Kim, Y.; Kim, D.J.; Grailhe, R.; Kim, Y.K. Bimolecular fluorescence complementation; lighting-up tau–tau interaction in living cells. PLoS ONE 2013, 8, e81682. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lim, S.; Haque, M.M.; Ryoo, N.; Hong, H.S.; Rhim, H.; Lee, D.-E.; Chang, Y.-T.; Lee, J.-S.; Cheong, E.; et al. Identification of disulfide cross-linked tau dimer responsible for tau propagation. Sci. Rep. 2015, 5, 15231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lim, S.; Haque, M.M.; Nam, G.; Ryoo, N.; Rhim, H.; Kim, Y.K. Monitoring of intracellular tau aggregation regulated by OGA/OGT inhibitors. Int. J. Mol. Sci. 2015, 16, 20212–20224. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Kim, D.; Ju, S.; Shin, S.; Cho, I.J.; Park, S.H.; Grailhe, R.; Lee, C.; Kim, Y.K. Glioblastoma-secreted soluble CD44 activates tau pathology in the brain. Exp. Mol. Med. 2018, 50, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carminati, J.L.; Stearns, T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 1997, 138, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Rusan, N.M.; Fagerstrom, C.J.; Yvon, A.-M.C.; Wadsworth, P. Cell cycle-dependent changes in microtubule dynamics in living cells expressing green fluorescent protein-α tubulin. Mol. Biol. Cell 2001, 12, 971–980. [Google Scholar] [CrossRef] [PubMed]
- Kellogg, E.H.; Hejab, N.M.; Poepsel, S.; Downing, K.H.; DiMaio, F.; Nogales, E. Near-atomic model of microtubule–tau interactions. Science 2018, 360, 1242–1246. [Google Scholar] [CrossRef] [PubMed]
- Snyder, J.P.; Nettles, J.H.; Cornett, B.; Downing, K.H.; Nogales, E. The binding conformation of Taxol in β-tubulin: A model based on electron crystallographic density. Proc. Natl. Acad. Sci. USA 2001, 98, 5312–5316. [Google Scholar] [CrossRef] [PubMed]
- Gupta, M.L.; Bode, C.J.; Georg, G.I.; Himes, R.H. Understanding tubulin–Taxol interactions: Mutations that impart Taxol binding to yeast tubulin. Proc. Natl. Acad. Sci. USA 2003, 100, 6394–6397. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Orr, G.A.; Chaudhary, A.G.; Kingston, D.G.; Horwitz, S.B. Characterization of the Taxol Binding Site on the Microtubule 2-(m-AZIDOBENZOYL) TAXOL PHOTOLABELS A PEPTIDE (AMINO ACIDS 217-231) of β-TUBULIN. J. Biol. Chem. 1995, 270, 20235–20238. [Google Scholar] [CrossRef] [PubMed]
- Amos, L.A.; Löwe, J. How Taxol® stabilises microtubule structure. Chem. Biol. 1999, 6, R65–R69. [Google Scholar] [CrossRef]
- Kar, S.; Fan, J.; Smith, M.J.; Goedert, M.; Amos, L.A. Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J. 2003, 22, 70–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makrides, V.; Massie, M.R.; Feinstein, S.C.; Lew, J. Evidence for two distinct binding sites for tau on microtubules. Proc. Natl. Acad. Sci. USA 2004, 101, 6746–6751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, G.C.; Timasheff, S.N. Thermodynamic linkage between tubulin self-association and the binding of vinblastine. Biochemistry 1980, 19, 1355–1365. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.S.; Wolff, J. Localization of the vinblastine-binding site on β-tubulin. J. Biol. Chem. 1996, 271, 14707–14711. [Google Scholar] [CrossRef] [PubMed]
- Rai, S.S.; Wolff, J. Dissociation of tubulin assembly-inhibiting and aggregation-promoting activities by a vinblastine derivative. FEBS Lett. 1997, 416, 251–253. [Google Scholar] [CrossRef] [Green Version]
- Vasquez, R.J.; Howell, B.; Yvon, A.; Wadsworth, P.; Cassimeris, L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell 1997, 8, 973–985. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Schwarz, P.M.; Ludueña, R.F. Interaction of nocodazole with tubulin isotypes. Drug Dev. Res. 2002, 55, 91–96. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Xiao, M.; Li, W.; Miller, D.D. An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm. Res. 2012, 29, 2943–2971. [Google Scholar] [CrossRef] [PubMed]
- Ho, Y.S.; Yang, X.; Lau, J.C.F.; Hung, C.H.L.; Wuwongse, S.; Zhang, Q.; Wang, J.; Baum, L.; So, K.-F.; Chang, R.C.-C. Endoplasmic reticulum stress induces tau pathology and forms a vicious cycle: Implication in Alzheimer’s disease pathogenesis. J. Alzheimer Dis. 2012, 28, 839–854. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.; Haque, M.M.; Su, D.; Kim, D.; Lee, J.S.; Chang, Y.T.; Kim, Y.K. Development of a BODIPY-based fluorescent probe for imaging pathological tau aggregates in live cells. Chem. Commun. 2017, 53, 1607–1610. [Google Scholar] [CrossRef] [PubMed]




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, S.; Lim, S.; Jeong, H.; Kwan, L.T.; Kim, Y.K. Visualization of Tau–Tubulin Interaction in a Living Cell Using Bifluorescence Complementation Technique. Int. J. Mol. Sci. 2018, 19, 2978. https://doi.org/10.3390/ijms19102978
Shin S, Lim S, Jeong H, Kwan LT, Kim YK. Visualization of Tau–Tubulin Interaction in a Living Cell Using Bifluorescence Complementation Technique. International Journal of Molecular Sciences. 2018; 19(10):2978. https://doi.org/10.3390/ijms19102978
Chicago/Turabian StyleShin, Seulgi, Sungsu Lim, Hyeanjeong Jeong, Li Ting Kwan, and Yun Kyung Kim. 2018. "Visualization of Tau–Tubulin Interaction in a Living Cell Using Bifluorescence Complementation Technique" International Journal of Molecular Sciences 19, no. 10: 2978. https://doi.org/10.3390/ijms19102978
APA StyleShin, S., Lim, S., Jeong, H., Kwan, L. T., & Kim, Y. K. (2018). Visualization of Tau–Tubulin Interaction in a Living Cell Using Bifluorescence Complementation Technique. International Journal of Molecular Sciences, 19(10), 2978. https://doi.org/10.3390/ijms19102978