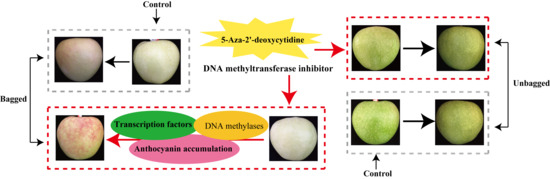
Transcriptome Profiling Reveals Transcriptional Regulation by DNA Methyltransferase Inhibitor 5-Aza-2′-Deoxycytidine Enhancing Red Pigmentation in Bagged “Granny Smith” Apples (Malus domestica)
Abstract
:1. Introduction
2. Results
2.1. Changes in Red Pigmentation Patterns in the Skin of “Granny Smith” Apples
2.2. RNA-Seq and Annotation Analysis of Differentially Expressed Genes (DEGs)
2.3. Expression of Anthocyanin Biosynthesis-Related Genes
2.4. Transcription Factors Are Differentially Expressed in Response to 5-Aza-dC Treatment
2.5. Expression of DNA Methylation-Related Genes
2.6. Quantitative Real-Time PCR (qRT-PCR) Validation of DEGs
3. Discussion
3.1. 5-Aza-dC Affects Anthocyanin Accumulation in the Skin of “Granny Smith” Apples
3.2. Epigenetic Regulation of MYB in “Granny Smith” Apples Following 5-Aza-dC Treatment
3.3. Other TFs Related to Anthocyanin Biosynthesis Following 5-Aza-dC Treatment
4. Materials and Methods
4.1. Plant Materials and Experimental Treatments
4.2. Measurement of Anthocyanin and Chlorophyll Content
4.3. RNA Extraction, Library Preparation, and RNA-Seq
4.4. RNA-Seq Data Processing and Mapping of Reads to the Apple Genome
4.5. RNA-Seq Data Analysis
4.6. RT-qPCR Validation
4.7. Data Accessibility
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Van Nocker, S.; Berry, G.; Najdowski, J.; Michelutti, R.; Luffman, M.; Forsline, P.; Alsmairat, N.; Beaudry, R.; Nair, M.G.; Ordidge, M. Genetic diversity of red-fleshed apples (Malus). Euphytica 2012, 185, 281–293. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Zheng, D.M.; Han, Y.P.; Khan, M.A.; Soria-Guerra, R.E.; Korban, S.S. Transcriptome analysis of the exocarp of apple fruit identifies light-induced genes involved in red color pigmentation. Gene 2014, 534, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.; Wu, X.; Liu, R.H. Antioxidant activity of apple peels. J. Agric. Food Chem. 2003, 51, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Weisel, T.; Baum, M.; Eisenbrand, G.; Dietrich, H.; Will, F.; Stockis, J.P.; Kulling, S.; Rüfer, C.; Johannes, C.; Janzowski, C. An anthocyanin/polyphenolic-rich fruit juice reduces oxidative DNA damage and increases glutathione level in healthy probands. Biotechnol. J. 2006, 1, 388–397. [Google Scholar] [CrossRef] [PubMed]
- He, J.A.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, J.E. Regulation of skin color in apples. Crit. Rev. Plant Sci. 1992, 10, 487–502. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, X.; Zhao, Z. Effects of fruit bagging on anthocyanins, sugars, organic acids, and color properties of ‘Granny Smith’ and ‘Golden Delicious’ during fruit maturation. Eur. Food Res. Technol. 2013, 236, 329–339. [Google Scholar] [CrossRef]
- Honda, C.; Kotoda, N.; Wada, M.; Kondo, S.; Kobayashi, S.; Soejima, J.; Zhang, Z.; Tsuda, T.; Moriguchi, T. Anthocyanin biosynthetic genes are coordinately expressed during red coloration in apple skin. Plant Physiol. Biotechnol. 2002, 40, 955–962. [Google Scholar] [CrossRef]
- Takos, A.M.; Jaffé, F.W.; Jacob, S.R.; Bogs, J.; Robinson, S.P.; Walker, A.R. Light-induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006, 142, 1216–1232. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, N.A.; Glover, B.J. MYB-bHLH-WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, N.; Liu, J.; Qu, C.; Wang, Y.; Jiang, S.; Lu, N.; Wang, D.; Zhang, Z.; Chen, X. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol. Biol. 2017, 94, 149–165. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD repeat (MBW) pigment regulatory model: Addition of a WRKY factor and co-option of an anthocyanin MYB for βlain regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Du, H.; Zhai, R.; Song, L.; Ma, F.; Xu, L. Transcriptome analysis reveals candidate genes related to color fading of ‘Red Bartlett’ (Pyrus communis L.). Front. Plant Sci. 2017, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.J.; Chuang, Y.N.; Chiou, C.Y.; Chin, D.C.; Shen, F.Q.; Yeh, K.W. Methylation effect on chalcone synthase gene expression determines anthocyanin pigmentation in floral tissues of two Oncidium orchid cultivars. Planta 2012, 236, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.G.; Meng, D.; Wang, A.D.; Li, T.L.; Jiang, S.L.; Cong, P.H.; Li, T.Z. The methylation of the PcMYB10 promoter is associated with green-skinned sport in max red Bartlett pear. Plant Physiol. 2013, 162, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Telias, A.; Kui, L.W.; Stevenson, D.E.; Cooney, J.M.; Hellens, R.P.; Allan, A.C.; Hoover, E.E.; Bradeen, J.M. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011, 11, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.Q.; Jing, C.J.; Chang, B.; Ya, J.Y.; Liang, B.W.; Liu, L.; Yang, Y.Z.; Zhao, Z.Y. The effect of promoter methylation on MdMYBl expression determines the level of anthocyanin accumulation in skins of two non-red apple cultivars. BMC Plant Biol. 2018, 18, 108. [Google Scholar] [CrossRef] [PubMed]
- Bai, S.; Tuan, P.A.; Saito, T.; Honda, C.; Hatsuyama, Y.; Ito, A.; Moriguchi, T. Epigenetic regulation of MdMYB1 is associated with paper bagging-induced red pigmentation of apples. Planta 2016, 244, 573–586. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.W.L.; Henderson, I.R.; Jacobsen, S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005, 6, 351. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhu, J.K. Active DNA demethylation in plants and animals. Cold Spring Harb. Symp. Quant. Biol. 2012, 77, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.D.; Wang, X.; Cao, H.B.; Xu, H.D.; Xu, Q.; Deng, X.X. Dynamic changes in methylome and transcriptome patterns in response to methyltransferase inhibitor 5-azacytidine treatment in citrus. DNA Res. 2017, 24, 509–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, W.; Li, B.; Wang, Q.; Wang, B.; Duan, X.; Ding, L.; Lu, Y.; Liu, L.W.; La, H. Analysis of the DNA methylation patterns and transcriptional regulation of the NB-LRR-encoding gene family in Arabidopsis thaliana. Plant Mol. Biol. 2018, 96, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Akimoto, K.; Katakami, H.; Kim, H.J.; Ogawa, E.; Sano, C.M.; Wada, Y.; Sano, H. Epigenetic inheritance in rice plants. Ann. Bot. 2007, 100, 205–217. [Google Scholar] [CrossRef] [PubMed]
- Burn, J.E.; Bagnall, D.J.; Metzger, J.D.; Dennis, E.S.; Peacock, W.J. DNA methylation, vernalization, and the initiation of flowering. Proc. Natl. Acad. Sci. USA 1993, 90, 287–291. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Tanino, K.K.; Horner, K.N.; Robinson, S.J. Quantitative trait variation is revealed in a novel hypomethylated population of woodland strawberry (Fragaria vesca). BMC Plant Biol. 2016, 16, 240. [Google Scholar] [CrossRef] [PubMed]
- Marfil, C.F.; Asurmendi, S.; Masuelli, R.W. Changes in micro RNA expression in a wild tuber-bearing Solanum species induced by 5-Azacytidine treatment. Plant Cell Rep. 2012, 31, 1449–1461. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Zhang, X.J.; Liu, Y.L.; Shi, X.Y.; Wang, Y.J.; Zhang, C.F.; Zhao, Z.Y. The effect of fruit bagging on the color, phenolic compounds and expression of the anthocyanin biosynthetic and regulatory genes on the ‘Granny Smith’ apples. Eur. Food Res. Technol. 2013, 237, 875–885. [Google Scholar] [CrossRef]
- Zhang, X.J.; Wang, L.X.; Liu, Y.L.; Chen, X.X.; Yang, Y.Z.; Zhao, Z.Y. Differential gene expression analysis of ‘Granny Smith’ apple (Malus domestica Borkh.) during fruit skin coloration. S. Afr. J. Bot. 2013, 88, 125–131. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.M.; Lloyd, A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espley, R.V.; Brendolise, C.; Chagné, D.; Kuttyamma, S.; Green, S.; Volz, R.; Putterill, J.; Schouten, H.J.; Gardiner, S.E.; Hellens, R.P. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell 2009, 21, 168–183. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.T.; Feng, S.Q.; Jiao, Q.Q.; Liu, C.C.; Zhang, W.W.; Chen, W.Y.; Chen, X.S. Comparison of MdMYB1 sequences and expression of anthocyanin biosynthetic and regulatory genes between Malus domestica Borkh. cultivar ‘Ralls’ and its blushed sport. Euphytica 2012, 185, 157–170. [Google Scholar] [CrossRef]
- Ravaglia, D.; Espley, R.V.; Henrykirk, R.A.; Andreotti, C.; Ziosi, V.; Hellens, R.P.; Costa, G.; Allan, A.C. Transcriptional regulation of flavonoid biosynthesis in nectarine (Prunus persica) by a set of R2R3 MYB transcription factors. BMC Plant Biol. 2013, 13, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, Y.H.; Yan, O.Z.; Tapken, W.; Kim, H.J.; Lavanway, A.M.; Howard, L.; Pilon, M.; Kieber, J.J.; Schaller, G.E. Functional characterization of the GATA transcription factors GNC and CGA1 reveals their key role in chloroplast development, growth, and division in Arabidopsis. Plant Physiol. 2012, 160, 332–348. [Google Scholar] [CrossRef] [PubMed]
- Morishita, T.; Kojima, Y.; Maruta, T.; Nishizawa-Yokoi, A.; Yabuta, Y.; Shigeoka, S. Arabidopsis NAC transcription factor, ANAC078, regulates flavonoid biosynthesis under high-light. Plant Cell Physiol. 2009, 50, 2210–2222. [Google Scholar] [CrossRef] [PubMed]
- Qian, M.J.; Ni, J.B.; Niu, Q.F.; Bai, S.L.; Bao, L.; Li, J.Z.; Sun, Y.W.; Zhang, D.; Teng, Y.W. Response of miR156-SPL module during the red peel coloration of bagging-treated Chinese sand pear (Pyrus pyrifolia Nakai). Front. Physiol. 2017, 8, 550. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Ratcliffe, O.J. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000, 3, 423–434. [Google Scholar] [CrossRef]
- Amato, A.; Cavallini, E.; Zenoni, S.; Finezzo, L.; Begheldo, M.; Ruperti, B.; Tornielli, G.B. A grapevine TTG2-like WRKY transcription factor is involved in regulating vacuolar transport and flavonoid biosynthesis. Front. Plant Sci. 2017, 7, 1979. [Google Scholar] [CrossRef] [PubMed]
- Qu, D.; Yan, F.; Meng, R.; Jiang, X.B.; Yang, H.J.; Gao, Z.Y.; Dong, Y.H.; Yang, Y.Z.; Zhao, Z.Y. Identification of microRNAs and their targets associated with fruit-bagging and subsequent sunlight re-exposure in the “Granny Smith” apple exocarp using high-throughput sequencing. Front. Plant Sci. 2016, 7, 27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Che, F.; Wang, L.; Meng, R.; Zhang, X.; Zhao, Z. Fruit coloration and anthocyanin biosynthesis after bag removal in non-red and red apples (Malus × domestica Borkh.). Molecules 2013, 18, 1549–1563. [Google Scholar] [CrossRef] [PubMed]
- Lichtenthaler, H.K.; Wellburn, A.R. Determinations of total carotenoids and chlorophylls a and b of leaf extracts in different solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Velasco, R.; Zharkikh, A.; Affourtit, J.; Dhingra, A.; Cestaro, A.; Kalyanaraman, A.; Fontana, P.; Bhatnagar, S.K.; Troggio, M.; Pruss, D. The genome of the domesticated apple (Malus × domestica Borkh.). Nat. Genet. 2010, 42, 833–839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.Q.; Yu, C.; Li, Y.R.; Lam, T.W.; Yiu, S.M.; Kristiansen, K.; Wang, J. SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 2009, 25, 1966–1967. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 1995, 57, 289–300. [Google Scholar]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, D480–D484. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using realtime quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]







| Sample | Clean Reads | Mapped Reads (%) | Uniquely Mapped Reads (%) | Expressed Genes |
|---|---|---|---|---|
| CK1-1 | 290,141,72 | 72.19 | 63.60 | 31713 |
| CK1-2 | 250,432,60 | 70.62 | 62.39 | 29085 |
| CK2-1 | 328,395,82 | 55.89 | 50.16 | 23434 |
| CK2-2 | 295,493,66 | 64.48 | 57.98 | 26639 |
| T1-1 | 238,889,26 | 70.18 | 61.79 | 30322 |
| T1-2 | 270,839,02 | 71.39 | 62.91 | 31195 |
| T2-1 | 319,614,16 | 72.05 | 64.33 | 30597 |
| T2-2 | 315,852,92 | 71.33 | 63.55 | 30470 |
| Pathway | DEGs with Pathway Annotation (309) | All Genes with Pathway Annotation (10,222) | p-Value | Pathway ID |
|---|---|---|---|---|
| Flavonoid biosynthesis | 12 (3.88%) | 95 (0.93%) | 0.000027 | ko00941 |
| Glutathione metabolism | 17 (5.5%) | 212 (2.07%) | 0.000235 | ko00480 |
| Phenylpropanoid biosynthesis | 25 (8.09%) | 393 (3.84%) | 0.000355 | ko00940 |
| Pyruvate metabolism | 15 (4.85%) | 210 (2.05%) | 0.001767 | ko00620 |
| Phenylalanine metabolism | 8 (2.59%) | 80 (0.78%) | 0.002791 | ko00360 |
| Aminoacyl-tRNA biosynthesis | 11 (3.56%) | 144 (1.41%) | 0.004247 | ko00970 |
| Linoleic acid metabolism | 5 (1.62%) | 41 (0.4%) | 0.007505 | ko00591 |
| Peroxisome | 13 (4.21%) | 200 (1.96%) | 0.007693 | ko04146 |
| Fatty acid elongation | 6 (1.94%) | 60 (0.59%) | 0.009244 | ko00062 |
| Carbon metabolism | 28 (9.06%) | 582 (5.69%) | 0.009922 | ko01200 |
| Monoterpenoid biosynthesis | 4 (1.29%) | 30 (0.29%) | 0.012073 | ko00902 |
| Nitrogen metabolism | 6 (1.94%) | 69 (0.68%) | 0.017687 | ko00910 |
| Butanoate metabolism | 4 (1.29%) | 34 (0.33%) | 0.018598 | ko00650 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, C.; Liang, B.; Chang, B.; Liu, L.; Yan, J.; Yang, Y.; Zhao, Z. Transcriptome Profiling Reveals Transcriptional Regulation by DNA Methyltransferase Inhibitor 5-Aza-2′-Deoxycytidine Enhancing Red Pigmentation in Bagged “Granny Smith” Apples (Malus domestica). Int. J. Mol. Sci. 2018, 19, 3133. https://doi.org/10.3390/ijms19103133
Ma C, Liang B, Chang B, Liu L, Yan J, Yang Y, Zhao Z. Transcriptome Profiling Reveals Transcriptional Regulation by DNA Methyltransferase Inhibitor 5-Aza-2′-Deoxycytidine Enhancing Red Pigmentation in Bagged “Granny Smith” Apples (Malus domestica). International Journal of Molecular Sciences. 2018; 19(10):3133. https://doi.org/10.3390/ijms19103133
Chicago/Turabian StyleMa, Changqing, Bowen Liang, Bo Chang, Li Liu, Jiuying Yan, Yazhou Yang, and Zhengyang Zhao. 2018. "Transcriptome Profiling Reveals Transcriptional Regulation by DNA Methyltransferase Inhibitor 5-Aza-2′-Deoxycytidine Enhancing Red Pigmentation in Bagged “Granny Smith” Apples (Malus domestica)" International Journal of Molecular Sciences 19, no. 10: 3133. https://doi.org/10.3390/ijms19103133
APA StyleMa, C., Liang, B., Chang, B., Liu, L., Yan, J., Yang, Y., & Zhao, Z. (2018). Transcriptome Profiling Reveals Transcriptional Regulation by DNA Methyltransferase Inhibitor 5-Aza-2′-Deoxycytidine Enhancing Red Pigmentation in Bagged “Granny Smith” Apples (Malus domestica). International Journal of Molecular Sciences, 19(10), 3133. https://doi.org/10.3390/ijms19103133