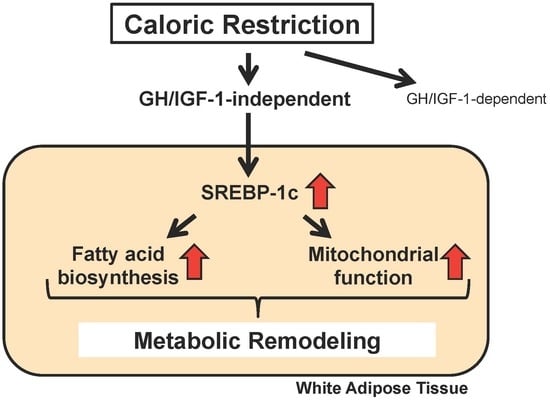
SREBP-1c-Dependent Metabolic Remodeling of White Adipose Tissue by Caloric Restriction
Abstract
:1. Introduction
2. Identification of SREBP-1c as a Novel Mediator of CR
3. Another SREBP-1c-Dependent Mechanism in the Effects of CR: Mitochondrial Activation
4. Proposed Mechanisms for the CR-Induced Upregulation of SREBP-1c
5. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Weindruch, R.; Walford, R.L. Retardation of Aging and Disease by Dietary Restriction; Charles C Thomas: Springfield, IL, USA, 1988. [Google Scholar]
- Yu, B.P. Modulation of Aging Processes by Dietary Restriction; CRC Press: Boca Raton, FL, USA, 1994. [Google Scholar]
- Lane, M.A.; Black, A.; Ingram, D.K.; Roth, G.S. Calorie restriction in non-human primates: Implications for age-related disease risk. J. Anti-Aging Med. 1998, 1, 315–326. [Google Scholar] [CrossRef]
- Roth, G.S.; Lane, M.A.; Ingram, D.K.; Mattison, J.A.; Elahi, D.; Tobin, J.D.; Muller, D.; Metter, E.J. Biomarkers of caloric restriction may predict longevity in humans. Science 2002, 297, 811. [Google Scholar] [CrossRef] [PubMed]
- Mattison, J.A.; Colman, R.J.; Beasley, T.M.; Allison, D.B.; Kemnitz, J.W.; Roth, G.S.; Ingram, D.K.; Weindruch, R.; de Cabo, R.; Anderson, R.M. Caloric restriction improves health and survival of rhesus monkeys. Nat. Commun. 2017, 8, 14063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chung, K.W.; Kim, D.H.; Park, M.H.; Choi, Y.J.; Kim, N.D.; Lee, J.; Yu, B.P.; Chung, H.Y. Recent advances in calorie restriction research on aging. Exp. Gerontol. 2013, 48, 1049–1053. [Google Scholar] [CrossRef] [PubMed]
- Guarente, L. Calorie restriction and sirtuins revisited. Genes Dev. 2013, 27, 2072–2085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Li, S.; Yang, L.; Huang, P.; Li, W.; Wang, S.; Zhao, G.; Zhang, M.; Pang, X.; Yan, Z.; et al. Structural modulation of gut microbiota in life-long calorie-restricted mice. Nat. Commun. 2013, 4, 2163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, S.J.; Madrigal-Matute, J.; Scheibye-Knudsen, M.; Fang, E.; Aon, M.; González-Reyes, J.A.; Cortassa, S.; Kaushik, S.; Gonzalez-Freire, M.; Patel, B.; et al. Effects of Sex, Strain, and Energy Intake on Hallmarks of Aging in Mice. Cell Metab. 2016, 23, 1093–1112. [Google Scholar] [CrossRef] [PubMed]
- Gnacińska, M.; Małgorzewicz, S.; Stojek, M.; Łysiak-Szydłowska, W.; Sworczak, K. Role of adipokines in complications related to obesity: A review. Adv. Med. Sci. 2009, 54, 150–157. [Google Scholar] [CrossRef] [PubMed]
- Torres-Leal, F.L.; Fonseca-Alaniz, M.H.; Rogero, M.M.; Tirapegui, J. The role of inflamed adipose tissue in the insulin resistance. Cell Biochem. Funct. 2010, 28, 623–631. [Google Scholar] [CrossRef] [PubMed]
- DeClercq, V.; Taylor, C.; Zahradka, P. Adipose tissue: The link between obesity and cardiovascular disease. Cardiovasc. Hematol. Disord. Drug Targets 2008, 8, 228–237. [Google Scholar] [CrossRef] [PubMed]
- Frayn, K.N. Adipose tissue as a buffer for daily lipid flux. Diabetologia 2002, 45, 1201–1210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blüher, M.; Kahn, B.B.; Kahn, C.R. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 2003, 299, 572–574. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.H.; Lin, W.D.; Huang, S.Y.; Lee, Y.H. Effect of a C/EBP gene replacement on mitochondrial biogenesis in fat cells. Genes Dev. 2004, 18, 1970–1975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argmann, C.; Dobrin, R.; Heikkinen, S.; Auburtin, A.; Pouilly, L.; Cock, T.A.; Koutnikova, H.; Zhu, J.; Schadt, E.E.; Auwerx, J. Pparγ2 is a key driver of longevity in the mouse. PLoS Genet. 2009, 5, e1000752. [Google Scholar] [CrossRef] [PubMed]
- Otabe, S.; Yuan, X.; Fukutani, T.; Wada, N.; Hashinaga, T.; Nakayama, H.; Hirota, N.; Kojima, M.; Yamada, K. Overexpression of human adiponectin in transgenic mice results in suppression of fat accumulation and prevention of premature death by high-calorie diet. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E210–E218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaza, H.; Komatsu, T.; To, K.; Toyama, H.; Chiba, T.; Higami, Y.; Shimokawa, I. Involvement of insulin-like growth factor-1 in the effect of caloric restriction: Regulation of plasma adiponectin and leptin. J. Gerontol. A Biol. Sci. Med. Sci. 2007, 62, 27–33. [Google Scholar] [CrossRef]
- Zhu, M.; Lee, G.D.; Ding, L.; Hu, J.; Qiu, G.; de Cabo, R.; Bernier, M.; Ingram, D.K.; Zou, S. Adipogenic signaling in rat white adipose tissue: Modulation by aging and calorie restriction. Exp. Gerontol. 2007, 42, 733–744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okita, N.; Hayashida, Y.; Kojima, Y.; Fukushima, M.; Yuguchi, K.; Mikami, K.; Yamauchi, A.; Watanabe, K.; Noguchi, M.; Nakamura, M.; et al. Differential responses of white adipose tissue and brown adipose tissue to caloric restriction in rats. Mech. Ageing Dev. 2012, 133, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, D.A. Toward a unified theory of caloric restriction and longevity regulation. Mech. Ageing Dev. 2005, 126, 987–1002. [Google Scholar] [CrossRef] [PubMed]
- Al-Regaiey, K.A.; Masternak, M.M.; Bonkowski, M.; Sun, L.; Bartke, A. Long-lived growth hormone receptor knockout mice: Interaction of reduced insulin-like growth factor I/insulin signaling and caloric restriction. Endocrinology 2005, 146, 851–860. [Google Scholar] [CrossRef] [PubMed]
- Bonkowski, M.S.; Rocha, J.S.; Masternak, M.M.; Al Regaiey, K.A.; Bartke, A. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc. Natl. Acad. Sci. USA 2006, 103, 7901–7905. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef]
- Hesp, K.; Smant, G.; Kammenga, J.E. Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age related disease. Exp. Gerontol. 2015, 72, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Yamaza, H.; Komatsu, T.; Wakita, S.; Kijogi, C.; Park, S.; Hayashi, H.; Chiba, T.; Mori, R.; Furuyama, T.; Mori, N.; et al. FoxO1 is involved in the antineoplastic effect of calorie restriction. Aging Cell 2010, 9, 372–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimokawa, I.; Komatsu, T.; Hayashi, N.; Kim, S.E.; Kawata, T.; Park, S.; Hayashi, H.; Yamaza, H.; Chiba, T.; Mori, R. The life-extending effect of dietary restriction requires Foxo3 in mice. Aging Cell 2015, 14, 707–709. [Google Scholar] [CrossRef] [PubMed]
- Bartke, A.; Wright, J.C.; Mattison, J.A.; Ingram, D.K.; Miller, R.A.; Roth, G.S. Extending the lifespan of long-lived mice. Nature 2001, 414, 412. [Google Scholar] [CrossRef] [PubMed]
- Shimokawa, I.; Higami, Y.; Tsuchiya, T.; Otani, H.; Komatsu, T.; Chiba, T.; Yamaza, H. Life span extension by reduction of the growth hormone-insulin-like growth factor-1 axis: Relation to caloric restriction. FASEB J. 2003, 17, 1108–1109. [Google Scholar] [CrossRef] [PubMed]
- Higami, Y.; Tsuchiya, T.; Chiba, T.; Yamaza, H.; Muraoka, I.; Hirose, M.; Komatsu, T.; Shimokawa, I. Hepatic gene expression profile of lipid metabolism in rats: Impact of caloric restriction and growth hormone/insulin-like growth factor-1 suppression. J. Gerontol. A Biol. Sci. Med. Sci. 2006, 61, 1099–1110. [Google Scholar] [CrossRef]
- Chujo, Y.; Fujii, N.; Okita, N.; Konishi, T.; Narita, T.; Yamada, A.; Haruyama, Y.; Tashiro, K.; Chiba, T.; Shimokawa, I.; et al. Caloric restriction-associated remodeling of rat white adipose tissue: Effects on the growth hormone/insulin-like growth factor-1 axis, sterol regulatory element binding protein-1, and macrophage infiltration. Age 2013, 35, 1143–1156. [Google Scholar] [CrossRef] [PubMed]
- Linford, N.J.; Beyer, R.P.; Gollahon, K.; Krajcik, R.A.; Malloy, V.L.; Demas, V.; Burmer, G.C.; Rabinovitch, P.S. Transcriptional response to aging and caloric restriction in heart and adipose tissue. Aging Cell 2007, 6, 673–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valle, A.; Sastre-Serra, J.; Roca, P.; Oliver, J. Modulation of white adipose tissue proteome by aging and calorie restriction. Aging Cell 2010, 9, 882–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osborne, T.F. Sterol regulatory element-binding proteins (SREBPs): Key regulators of nutritional homeostasis and insulin action. J. Biol. Chem. 2000, 275, 32379–32382. [Google Scholar] [CrossRef] [PubMed]
- Osborne, T.F.; Espenshade, P.J. Evolutionary conservation and adaptation in the mechanism that regulates SREBP action: What a long, strange tRIP it’s been. Genes Dev. 2009, 23, 2578–2591. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Shah, N.A.; Warrington, J.A.; Anderson, N.N.; Park, S.W.; Brown, M.S.; Goldstein, J.L. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc. Natl. Acad. Sci. USA 2003, 100, 12027–12032. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimano, H.; Horton, J.D.; Shimomura, I.; Hammer, R.E.; Brown, M.S.; Goldstein, J.L. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J. Clin. Invest. 1997, 99, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, I.; Hammer, R.E.; Richardson, J.A.; Ikemoto, S.; Bashmakov, Y.; Goldstein, J.L.; Brown, M.S. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: Model for congenital generalized lipodystrophy. Genes Dev. 1998, 12, 3182–3194. [Google Scholar] [CrossRef] [PubMed]
- Fujii, N.; Narita, T.; Okita, N.; Kobayashi, M.; Furuta, Y.; Chujo, Y.; Sakai, M.; Yamada, A.; Takeda, K.; Konishi, T.; et al. Sterol regulatory element-binding protein-1c orchestrates metabolic remodeling of white adipose tissue by caloric restriction. Aging Cell 2017, 16, 508–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.J.; Kaeberlein, M.; Andalis, A.A.; Sturtz, L.A.; Defossez, P.A.; Culotta, V.C.; Fink, G.R.; Guarente, L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002, 418, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Zid, B.M.; Rogers, A.N.; Katewa, S.D.; Vargas, M.A.; Kolipinski, M.C.; Lu, T.A.; Benzer, S.; Kapahi, P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 2009, 139, 149–160. [Google Scholar] [CrossRef] [PubMed]
- Nisoli, E.; Tonello, C.; Cardile, A.; Cozzi, V.; Bracale, R.; Tedesco, L.; Falcone, S.; Valerio, A.; Cantoni, O.; Clementi, E. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 2005, 310, 314–317. [Google Scholar] [CrossRef] [PubMed]
- Sreekumar, R.; Unnikrishnan, J.; Fu, A.; Nygren, J.; Short, K.R.; Schimke, J.; Barazzoni, R.; Nair, K.S. Effects of caloric restriction on mitochondrial function and gene transcripts in rat muscle. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E38–E43. [Google Scholar] [CrossRef] [PubMed]
- Civitarese, A.E.; Carling, S.; Heilbronn, L.K.; Hulver, M.H.; Ukropcova, B.; Deutsch, W.A.; Smith, S.R.; Ravussin, E. CALERIE Pennington Team. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007, 4, e76. [Google Scholar] [CrossRef] [PubMed]
- López-Lluch, G.; Hunt, N.; Jones, B.; Zhu, M.; Jamieson, H.; Hilmer, S.; Cascajo, M.V.; Allard, J.; Ingram, D.K.; Navas, P.; de Cabo, R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc. Natl. Acad. Sci. USA 2006, 103, 1768–1773. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lanza, I.R.; Zabielski, P.; Klaus, K.A.; Morse, D.M.; Heppelmann, C.J.; Bergen, H.R., 3rd; Dasari, S.; Walrand, S.; Short, K.R.; Johnson, M.L.; Robinson, M.M.; et al. Chronic caloric restriction preserves mitochondrial function in senescence without increasing mitochondrial biogenesis. Cell Metab. 2012, 16, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Spiegelman, B.M. Peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α): Transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003, 24, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Puigserver, P.; Wu, Z.; Park, C.W.; Graves, R.; Wright, M.; Spiegelman, B.M. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell 1998, 92, 829–839. [Google Scholar] [CrossRef]
- Wu, Z.; Puigserver, P.; Andersson, U.; Zhang, C.; Adelmant, G.; Mootha, V.; Troy, A.; Cinti, S.; Lowell, B.B.; Scarpulla, R.C.; et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999, 98, 115–124. [Google Scholar] [CrossRef]
- Evans, M.J.; Scarpulla, R.C. NRF-1: A trans-activator of nuclearencoded respiratory genes in animal cells. Genes Dev. 1990, 4, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Virbasius, J.V.; Virbasius, C.A.; Scarpulla, R.C. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 1993, 7, 380–392. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.M.; Barger, J.L.; Edwards, M.G.; Braun, K.H.; O’Connor, C.E.; Prolla, T.A.; Weindruch, R. Dynamic regulation of PGC-1α localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell 2008, 7, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Nemoto, S.; Fergusson, M.M.; Finkel, T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1α. J. Biol. Chem. 2005, 280, 16456–16460. [Google Scholar] [CrossRef] [PubMed]
- Rodgers, J.T.; Lerin, C.; Haas, W.; Gygi, S.P.; Spiegelman, B.M.; Puigserver, P. Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 2005, 434, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Ke, S.F.; Zhou, C.C.; Zhang, S.L.; Guan, Y.F.; Xu, T.Y.; Sheng, C.Q.; Wang, P.; Miao, C.Y. Nicotinamide phosphoribosyltransferase is required for the calorie restriction-mediated improvements in oxidative stress, mitochondrial biogenesis, and metabolic adaptation. J. Gerontol. A Biol. Sci. Med. Sci. 2014, 69, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Bulteau, A.L.; Ikeda-Saito, M.; Szweda, L.I. Redox-dependent modulation of aconitase activity in intact mitochondria. Biochemistry 2003, 42, 14846–14855. [Google Scholar] [CrossRef] [PubMed]
- Okita, N.; Tsuchiya, T.; Fukushima, M.; Itakura, K.; Yuguchi, K.; Narita, T.; Hashizume, Y.; Sudo, Y.; Chiba, T.; Shimokawa, I.; et al. Chronological analysis of caloric restriction-induced alteration of fatty acid biosynthesis in white adipose tissue of rats. Exp. Gerontol. 2015, 63, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Horton, J.D.; Shimomura, I.; Brown, M.S.; Hammer, R.E.; Goldstein, J.L.; Shimano, H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J. Clin. Invest. 1998, 101, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, M.; Yahagi, N.; Matsuzaka, T.; Takeuchi, Y.; Nakagawa, Y.; Takahashi, H.; Okazaki, H.; Iizuka, Y.; Ohashi, K.; Gotoda, T.; et al. SREBP-1-independent regulation of lipogenic gene expression in adipocytes. J. Lipid Res. 2007, 48, 1581–1591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soukas, A.; Cohen, P.; Socci, N.D.; Friedman, J.M. Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev. 2000, 14, 963–980. [Google Scholar] [PubMed]
- Stelmanska, E.; Korczynska, J.; Swierczynski, J. Tissue-specific effect of refeeding after short- and long-term caloric restriction on malic enzyme gene expression in rat tissues. Acta. Biochim. Pol. 2004, 51, 805–814. [Google Scholar] [PubMed]
- Shimomura, I.; Bashmakov, Y.; Horton, J.D. Increased levels of nuclear SREBP-1c associated with fatty livers in two mouse models of diabetes mellitus. J. Biol. Chem. 1999, 274, 30028–30032. [Google Scholar] [CrossRef] [PubMed]
- Shimomura, I.; Matsuda, M.; Hammer, R.E.; Bashmakov, Y.; Brown, M.S.; Goldstein, J.L. Decreased IRS-2 and increased SREBP-1c lead to mixed insulin resistance and sensitivity in livers of lipodystrophic and ob/ob mice. Mol. Cell 2000, 6, 77–86. [Google Scholar] [CrossRef]
- Hirschey, M.D.; Shimazu, T.; Jing, E.; Grueter, C.A.; Collins, A.M.; Aouizerat, B.; Stančáková, A.; Goetzman, E.; Lam, M.M.; Schwer, B.; et al. SIRT3 deficiency and mitochondrial protein hyperacetylation accelerate the development of the metabolic syndrome. Mol. Cell 2011, 44, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Liebman, S.E.; Lucia, M.S.; Phillips, C.L.; Levi, M. Calorie restriction modulates renal expression of sterol regulatory element binding proteins, lipid accumulation, and age-related renal disease. J. Am. Soc. Nephrol. 2005, 16, 2385–2394. [Google Scholar] [CrossRef] [PubMed]
- Suárez-Zamorano, N.; Fabbiano, S.; Chevalier, C.; Stojanović, O.; Colin, D.J.; Stevanović, A.; Veyrat-Durebex, C.; Tarallo, V.; Rigo, D.; Germain, S.; et al. Microbiota depletion promotes browning of white adipose tissue and reduces obesity. Nat. Med. 2015, 21, 1497–1501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geurts, L.; Neyrinck, A.M.; Delzenne, N.M.; Knauf, C.; Cani, P.D. Gut microbiota controls adipose tissue expansion, gut barrier and glucose metabolism: Novel insights into molecular targets and interventions using prebiotics. Benef. Microbes 2014, 5, 3–17. [Google Scholar] [CrossRef] [PubMed]
- Bleau, C.; Karelis, A.D.; St-Pierre, D.H.; Lamontagne, L. Crosstalk between intestinal microbiota, adipose tissue and skeletal muscle as an early event in systemic low-grade inflammation and the development of obesity and diabetes. Diabetes Metab. Res. Rev. 2015, 31, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Tang, Y.; Li, M.; Yang, P.; Liu, Z.; Yuan, J.; Zheng, P. Co-Administration of Cholesterol-Lowering Probiotics and Anthraquinone from Cassia obtusifolia L. Ameliorate Non-Alcoholic Fatty Liver. PLoS ONE 2015, 10, e0138078. [Google Scholar] [CrossRef] [PubMed]
- Alves, C.C.; Waitzberg, D.L.; de Andrade, L.S.; Dos Santos Aguiar, L.; Reis, M.B.; Guanabara, C.C.; Júnior, O.A.; Ribeiro, D.A.; Sala, P. Prebiotic and Synbiotic Modifications of Beta Oxidation and Lipogenic Gene Expression after Experimental Hypercholesterolemia in Rat Liver. Front Microbiol. 2017, 8, 2010. [Google Scholar] [CrossRef] [PubMed]
- Fraumene, C.; Manghina, V.; Cadoni, E.; Marongiu, F.; Abbondio, M.; Serra, M.; Palomba, A.; Tanca, A.; Laconi, E.; Uzzau, S. Caloric restriction promotes rapid expansion and long-lasting increase of Lactobacillus in the rat fecal microbiota. Gut Microbes 2018, 9, 104–114. [Google Scholar] [CrossRef] [PubMed]
- Fabbiano, S.; Suárez-Zamorano, N.; Chevalier, C.; Lazarević, V.; Kieser, S.; Rigo, D.; Leo, S.; Veyrat-Durebex, C.; Gaïa, N.; Maresca, M.; et al. Functional Gut Microbiota Remodeling Contributes to the Caloric Restriction-Induced Metabolic Improvements. Cell Metab. 2018, 24. [Google Scholar] [CrossRef] [PubMed]
- Bruss, M.D.; Khambatta, C.F.; Ruby, M.A.; Aggarwal, I.; Hellerstein, M.K. Calorie restriction increases fatty acid synthesis and whole body fat oxidation rates. Am. J. Physiol. Endocrinol. Metab. 2010, 298, E108–E116. [Google Scholar] [CrossRef] [PubMed]
- Narita, T.; Kobayashi, M.; Itakura, K.; Itagawa, R.; Kabaya, R.; Sudo, Y.; Okita, N.; Higami, Y. Differential response to caloric restriction of retroperitoneal, epididymal, and subcutaneous adipose tissue depots in rats. Exp. Gerontol. 2018, 104, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Akram, M. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem. Biophys. 2014, 68, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflugers Arch. 2004, 447, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Hiltunen, J.K.; Autio, K.J.; Schonauer, M.S.; Kursu, V.A.; Dieckmann, C.L.; Kastaniotis, A.J. Mitochondrial fatty acid synthesis and respiration. Biochim. Biophys. Acta 2010, 1797, 1195–1202. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kobayashi, M.; Fujii, N.; Narita, T.; Higami, Y. SREBP-1c-Dependent Metabolic Remodeling of White Adipose Tissue by Caloric Restriction. Int. J. Mol. Sci. 2018, 19, 3335. https://doi.org/10.3390/ijms19113335
Kobayashi M, Fujii N, Narita T, Higami Y. SREBP-1c-Dependent Metabolic Remodeling of White Adipose Tissue by Caloric Restriction. International Journal of Molecular Sciences. 2018; 19(11):3335. https://doi.org/10.3390/ijms19113335
Chicago/Turabian StyleKobayashi, Masaki, Namiki Fujii, Takumi Narita, and Yoshikazu Higami. 2018. "SREBP-1c-Dependent Metabolic Remodeling of White Adipose Tissue by Caloric Restriction" International Journal of Molecular Sciences 19, no. 11: 3335. https://doi.org/10.3390/ijms19113335
APA StyleKobayashi, M., Fujii, N., Narita, T., & Higami, Y. (2018). SREBP-1c-Dependent Metabolic Remodeling of White Adipose Tissue by Caloric Restriction. International Journal of Molecular Sciences, 19(11), 3335. https://doi.org/10.3390/ijms19113335