Nitrogen Limitation Alters the Response of Specific Genes to Biotic Stress
Abstract
:1. Introduction
2. Results
2.1. Effect of N Limitation on Gene Expression in Response to Erwinia amylovora (Ea)
2.2. N Availability Modulates Specific Defense-Related Genes
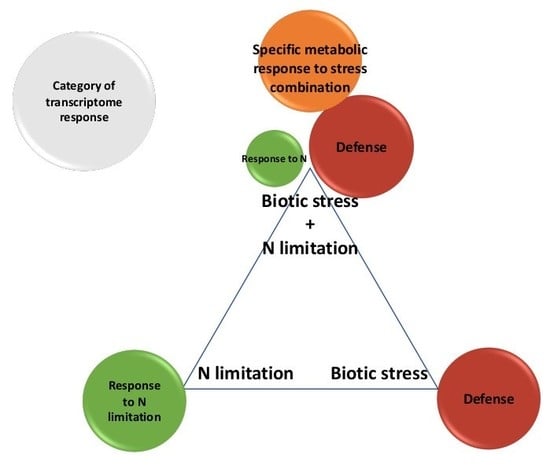
2.3. N Limitation and Biotic Stresses Interact
2.4. Defense-Associated Genes Are Prioritized in Response to the Combination of N Limitation with Different Pathogens
3. Discussion
4. Materials and Methods
4.1. Growth Conditions of Arabidopsis Plants
4.2. Pathogen Infections
4.3. RNA Isolation and qRT-PCR Analysis
4.4. Transcriptome Studies
4.5. Statistical Analysis of Microarray Data
4.6. Transcriptional Response Modes
4.7. Data Availability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| B | Bacteria |
| ET | Ethylene |
| GO | Gene Ontology |
| HPI | Hours post inoculation |
| JA | Jasmonic acid |
| MYB | MYeloBlastosis |
| N | Nitrogen |
| SA | Salicylic acid |
| TF | Transcription factor |
References
- Rasmussen, S.; Barah, P.; Suarez-Rodriguez, M.C.; Bressendorff, S.; Friis, P.; Costantino, P.; Bones, A.M.; Nielsen, H.B.; Mundy, J. Transcriptome Responses to Combinations of Stresses in Arabidopsis. Plant Physiol. 2013, 161, 1783–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, Y.H.; Halliday, K.J. A stress-free walk from Arabidopsis to crops. Curr. Opin. Biotechnol. 2011, 22, 281–286. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Rivero, R.M.; Shulaev, V.; Blumwald, E.; Mittler, R. Abiotic and biotic stress combinations. New Phytol. 2014, 203, 32–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, N.J.; Lilley, C.J.; Urwin, P.E. Identification of Genes Involved in the Response of Arabidopsis to Simultaneous Biotic and Abiotic Stresses. Plant Physiol. 2013, 162, 2028–2041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dordas, C. Role of nutrients in controlling plant diseases in sustainable agriculture. A review. Agron. Sustain. Dev. 2008, 28, 33–46. [Google Scholar] [CrossRef]
- Masclaux-Daubresse, C.; Daniel-Vedele, F.; Dechorgnat, J.; Chardon, F.; Gaufichon, L.; Suzuki, A. Nitrogen uptake, assimilation and remobilization in plants: Challenges for sustainable and productive agriculture. Ann. Bot. 2010, 105, 1141–1157. [Google Scholar] [CrossRef] [PubMed]
- Ballini, E.; Nguyen, T.T.; Morel, J.-B. Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice 2013, 6, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fagard, M.; Launay, A.; Clément, G.; Courtial, J.; Dellagi, A.; Farjad, M.; Krapp, A.; Soulié, M.C.; Masclaux-Daubresse, C. Nitrogen metabolism meets phytopathology. J. Exp. Bot. 2014, 65, 5643–5656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daugaard, H.; Sørensen, L.; Løschenkohl, B. Effect of Plant Spacing, Nitrogen Fertilisation, Post-Harvest Defoliation and Finger Harrowing in the Control of Botrytis cinerea Pers. in Strawberry. Eur. J. Hortic. Sci. 2016, 68, 77–82. [Google Scholar]
- Vega, A.; Canessa, P.; Hoppe, G.; Retamal, I.; Moyano, T.C.; Canales, J.; Gutiérrez, R.A.; Rubilar, J. Transcriptome analysis reveals regulatory networks underlying differential susceptibility to Botrytis cinerea in response to nitrogen availability in Solanum lycopersicum. Front. Plant Sci. 2015, 6, 1964. [Google Scholar] [CrossRef] [PubMed]
- Mur, L.A.J.; Simpson, C.; Kumari, A.; Gupta, A.K.; Gupta, K.J. Moving nitrogen to the centre of plant defence against pathogens. Ann. Bot. 2017, 119, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.L.; Forcat, S.; Beckmann, M.; Bennett, M.; Miller, S.J.; Baker, J.M.; Hawkins, N.D.; Vermeer, C.P.; Lu, C.; Lin, W.; et al. The metabolic transition during disease following infection of Arabidopsis thaliana by Pseudomonas syringae pv. tomato. Plant. J. 2010, 63, 443–457. [Google Scholar] [CrossRef] [PubMed]
- Snoeijers, S.; Pérez-García, A.; Joosten, M.; De Wit, P. The effect of nitrogen on disease development and gene expression in bacterial and fungal plant pathogens. Eur. J. Plant. Pathol. 2000, 106, 493. [Google Scholar] [CrossRef]
- Dietrich, C.R.; Ploß, K.; Heil, K. Constitutive and induced resistance to pathogens in Arabidopsis thaliana depends on nitrogen supply. Plant. Cell Environ. 2004, 27, 896–906. [Google Scholar] [CrossRef]
- Gupta, K.J.; Brotman, Y.; Segu, S.; Zeier, T.; Zeier, J.; Persijn, S.T.; Cristescu, S.M.; Harren, F.J.; Bauwe, H.; Fernie, A.R.; et al. The form of nitrogen nutrition affects resistance against Pseudomonas syringae pv. phaseolicola in tobacco. J. Exp. Bot. 2012, 64, 553–568. [Google Scholar] [CrossRef] [PubMed]
- Tiburcio, A.F.; Altabella, T.; Bitrián, M.; Alcázar, R. The roles of polyamines during the lifespan of plants: From development to stress. Planta 2014, 240, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Huot, B.; Yao, J.; Montgomery, B.L.; He, S.Y. Growth–Defense Tradeoffs in Plants: A Balancing Act to Optimize Fitness. Mol. Plant. 2014, 7, 1267–1287. [Google Scholar] [CrossRef] [PubMed]
- Peng, M.; Hudson, D.; Schofield, A.; Tsao, R.; Yang, R.; Gu, H.; Bi, Y.M.; Rothstein, S.J. Adaptation of Arabidopsis to nitrogen limitation involves induction of anthocyanin synthesis which is controlled by the NLA gene. J. Exp. Bot. 2008, 59, 2933–2944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lothier, J.; Gaufichon, L.; Sormani, R.; Lemaître, T.; Azzopardi, M.; Morin, H.; Chardon, F.; Reisdorf-Cren, M.; Avice, J.C.; Masclaux-Daubresse, C. The cytosolic glutamine synthetase GLN1;2 plays a role in the control of plant growth and ammonium homeostasis in Arabidopsis rosettes when nitrate supply is not limiting. J. Exp. Bot. 2011, 62, 1375–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreau, M.; Degrave, A.; Vedel, R.; Bitton, F.; Patrit, O.; Renou, J.P.; Barny, M.A.; Fagard, M. EDS1 contributes to nonhost resistance of Arabidopsis thaliana against Erwinia amylovora. Mol. Plant-Microbe Interact. 2012, 25, 421–430. [Google Scholar] [CrossRef] [PubMed]
- Ruepp, A. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 2004, 32, 5539–5545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alves, M.; Dadalto, S.; Gonçalves, A.; de Souza, G.; Barros, V.; Fietto, L. Transcription Factor Functional Protein-Protein Interactions in Plant Defense Responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Gou, M.; Liu, C.J. Arabidopsis Kelch Repeat F-Box Proteins Regulate Phenylpropanoid Biosynthesis via Controlling the Turnover of Phenylalanine Ammonia-Lyase. Plant Cell. 2014, 25, 4994–5010. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; He, M.; Zhu, Z.; Li, S.; Xu, Y.; Zhang, C.; Singer, S.D.; Wang, Y. Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotic and biotic stress. BMC Plant Biol. 2012, 12, 140. [Google Scholar] [CrossRef] [PubMed]
- Galon, Y.; Nave, R.; Boyce, J.M.; Nachmias, D.; Knight, M.R.; Fromm, H. Calmodulin-binding transcription activator (CAMTA) 3 mediates biotic defense responses in Arabidopsis. FEBS Lett. 2008, 582, 943–948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birkenbihl, R.P.; Liu, S.; Somssich, I.E. Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 2017, 38, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Camanes, G.; Pastor, V.; Cerezo, M.; García-Andrade, J.; Vicedo, B.; García-Agustín, P.; Flors, V. A deletion in NRT2.1 attenuates Pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 2012, 158, 1054–1066. [Google Scholar] [CrossRef] [PubMed]
- Dechorgnat, J.; Patrit, O.; Krapp, A.; Fagard, M.; Daniel-Vedele, F. Characterization of the Nrt2.6 Gene in Arabidopsis thaliana: A Link with Plant Response to Biotic and Abiotic Stress. PLoS ONE 2012, 7, e42491. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Ji, Y.; Bhuiyan, N.H.; Pilot, G.; Selvaraj, G.; Zou, J.; Wei, Y. Amino Acid Homeostasis Modulates Salicylic Acid-Associated Redox Status and Defense Responses in Arabidopsis. Plant Cell 2010, 22, 3845–3863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastor, V.; Gamir, J.; Camañes, G.; Cerezo, M.; Sanchez-Bel, P.; Flors, V. Disruption of the ammonium transporter AMT1.1 alters basal defenses generating resistance against Pseudomonas syringae and Plectosphaerella cucumerina. Front. Plant Sci. 2014, 5, 838. [Google Scholar] [CrossRef] [PubMed]
- Titarenko, E.; Rojo, E.; Leon, J.; Sanchez-Serrano, J.J. Jasmonic acid-dependent and -independent signaling pathways control wound-induced gene activation in Arabidopsis thaliana. Plant Physiol. 1997, 115, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Walters, D.R.; Bingham, I.J. Influence of nutrition on disease development caused by fungal pathogens: Implications for plant disease control. Ann. Appl. Biol. 2007, 151, 307–324. [Google Scholar] [CrossRef]
- Massad, T.J.; Dyer, L.A.; Vega, C.G. Costs of defense and a test of the carbon-nutrient balance and growth-differentiation balance hypotheses for two co-occurring classes of plant defense. PLoS ONE 2012, 7, e47554. [Google Scholar] [CrossRef] [PubMed]
- Coolen, S.; Proietti, S.; Hickman, R.; Davila Olivas, N.H.; Huang, P.P.; Van Verk, M.C.; Van Pelt, J.A.; Wittenberg, A.H.; De Vos, M.; Prins, M.; et al. Transcriptome dynamics of Arabidopsis during sequential biotic and abiotic stresses. Plant. J. 2016, 86, 249–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed]
- Degrave, A.; Fagard, M.; Perino, C.; Brisset, M.N.; Gaubert, S.; Laroche, S.; Patrit, O.; Barny, M.A. Erwinia amylovora type three–secreted proteins trigger cell death and defense responses in Arabidopsis thaliana. Mol. Plant Microb. Interact. 2008, 21, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Venisse, J.-S.; Malnoy, M.; Faize, M.; Paulin, J.-P.; Brisset, M.-N. Modulation of defense responses of Malus spp. during compatible and incompatible interactions with Erwinia amylovora. Mol. Plant-Microbe Interact. 2002, 15, 1204–1212. [Google Scholar] [CrossRef] [PubMed]
- De Bernonville, T.D.; Gaucher, M.; Flors, V.; Gaillard, S.; Paulin, J.P.; Dat, J.F.; Brisset, M.N. T3SS-dependent differential modulations of the jasmonic acid pathway in susceptible and resistant genotypes of Malus spp. challenged with Erwinia amylovora. Plant Sci. 2012, 188–189, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eulgem, T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 2005, 10, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.; Xie, Z.; Chen, W.; Glazebrook, J.; Chang, H.S.; Han, B.; Zhu, T.; Zou, G.; Katagiri, F. Quantitative Nature of Arabidopsis Responses during Compatible and Incompatible Interactions with the Bacterial Pathogen Pseudomonas syringae. Plant Cell 2003, 15, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Larionov, A.; Krause, A.; Miller, W. A standard curve based method for relative real time PCR data processing. BMC Bioinform. 2005, 6, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceccato, L.; Masiero, S.; Roy, D.S.; Bencivenga, S.; Roig-Villanova, I.; Ditengou, F.A.; Palme, K.; Simon, R.; Colombo, L. Maternal control of PIN1 is required for female gametophyte development in Arabidopsis. PLoS ONE 2013, 8, e66148. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miranda, K.M.; Espey, M.G.; Wink, D.A. A Rapid, Simple Spectrophotometric Method for Simultaneous Detection of Nitrate and Nitrite. Nitric Oxide-Biol. Chem. 2001, 5, 62–71. [Google Scholar] [CrossRef] [PubMed]



| Gene Number | Name | Function | Low N | Full N | ||
|---|---|---|---|---|---|---|
| Ea vs. Mock | Ea vs. Mock | |||||
| SA biosynthesis and responsive genes | ||||||
| AT1G74710 | ICS1 | BS | 6.14 | b | 5.73 | b |
| AT3G52430 | PAD4 | BS | 5.15 | b | 4.51 | b |
| AT3G48090 | EDS1 | S | 3.85 | b | 3.82 | b |
| AT4G39030 | EDS5 | S | 6.33 | b | 5.67 | b |
| AT1G64280 | NPR1 | S | 2.14 | a | 1.66 | a |
| AT2G14610 | PR1 | R | 5.10 | b | 3.23 | b |
| AT3G57260 | PR2 | R | 1.50 | a | 0.03 | |
| AT1G75040 | PR5 | R | 2.00 | b | 0.88 | |
| ET biosynthesis and responsive genes | ||||||
| AT1G05010 | ACO | BS | 2.64 | b | 1.62 | a |
| AT3G04580 | EIN4 | R | 2.76 | b | 2.66 | b |
| AT4G17500 | ERF-1 | R | 1.93 | a | 1.92 | a |
| AT2G40940 | ERS1 | R | 1.42 | a | 1.05 | a |
| AT1G66340 | ETR1 | R | 1.19 | 1.14 | ||
| JA biosynthesis and responsive genes | ||||||
| AT3G25770 | AOC2 | BS | −1.17 | −2.12 | b | |
| AT5G42650 | AOS | BS | −1.13 | −2.13 | b | |
| AT2G46370 | JAR1 | BS | −0.23 | −0.51 | ||
| AT3G45140 | LOX2 | BS | −0.35 | −0.97 | ||
| AT2G06050 | OPR3 | BS | 0.16 | 0.26 | ||
| AT1G19640 | JMT | R | −0.60 | −0.23 | ||
| AT3G16470 | JR1 | R | 0.08 | −2.01 | a | |
| AT2G26020 | PDF1.2b | R | −1.77 | a | −2.06 | a |
| AT3G12500 | PR3 | R | 0.06 | −0.06 | ||
| AT3G04720 | PR4 | R | 2.43 | b | 0.35 | |
| AT5G24770 | VSP2 | R | 0.14 | −0.02 | ||
| AT1G32640 | ATMYC2 | S | −1.70 | a | −1.82 | a |
| AT2G39940 | COI1 | S | 0.25 | 0.10 | ||
| Gene Name | Gene Number | N | B | N + B | Response Mode |
|---|---|---|---|---|---|
| Response to salicylic acid | |||||
| PR5 | AT1G75040 | −2.07 | 0.88 | −0.06 | cancelled |
| EDS5 | AT4G39030 | −1.09 | 5.67 | 5.24 | prioritized |
| PR1 | AT2G14610 | −2.71 | 3.23 | 2.40 | prioritized |
| ET/JA response and biosynthesis | |||||
| JR1 | AT3G16470 | −0.60 | −2.01 | −0.52 | cancelled |
| ETR1 | AT1G66340 | −0.41 | 1.14 | 0.78 | cancelled |
| LOX3 | AT1G17420 | −1.15 | 2.44 | 1.96 | prioritized |
| EDS1 pathway | |||||
| FMO1 | AT1G19250 | −1.28 | 5.85 | 5.02 | prioritized |
| AtNUDT7 | AT4G12720 | −1.10 | 3.28 | 2.97 | prioritized |
| WRKY transcription factors | |||||
| WRKY75 | AT5G13080 | −1.12 | 4.20 | 3.72 | prioritized |
| WRKY51 | AT5G64810 | −1.50 | 5.93 | 5.47 | prioritized |
| WRKY38 | AT5G22570 | −1.93 | 3.68 | 3.12 | prioritized |
| WRKY25 | AT2G30250 | −1.23 | 2.53 | 1.96 | prioritized |
| WRKY54 | AT2G40750 | −1.04 | 2.22 | 1.88 | prioritized |
| WRKY70 | AT3G56400 | −1.30 | 1.69 | 1.46 | prioritized |
| WRKY53 | AT4G23810 | −1.42 | 2.24 | 1.22 | prioritized |
| WRKY60 | AT2G25000 | −1.15 | 1.77 | 1.34 | prioritized |
| WRKY50 | AT5G26170 | −1.02 | 3.45 | 2.64 | prioritized |
| WRKY30 | AT5G24110 | −1.15 | 6.11 | 4.90 | prioritized |
| WRKY33 | AT2G38470 | −1.43 | 4.63 | 3.79 | prioritized |
| Resistance genes | |||||
| “LRR family protein” | AT5G45510 | −1.24 | 1.87 | 1.43 | prioritized |
| N metabolism | |||||
| GLN1.3 | AT3G17820 | −0.38 | −1.02 | −0.90 | cancelled |
| GDH3 | AT3G03910 | −0.06 | 1.21 | 0.24 | cancelled |
| AMT1.1 | AT4G13510 | −1.13 | 2.40 | 1.98 | prioritized |
| WR3 | AT5G50200 | −1.28 | 3.38 | 2.63 | prioritized |
| LHT1 | AT5G40780 | −1.30 | 3.04 | 2.72 | prioritized |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Farjad, M.; Rigault, M.; Pateyron, S.; Martin-Magniette, M.-L.; Krapp, A.; Meyer, C.; Fagard, M. Nitrogen Limitation Alters the Response of Specific Genes to Biotic Stress. Int. J. Mol. Sci. 2018, 19, 3364. https://doi.org/10.3390/ijms19113364
Farjad M, Rigault M, Pateyron S, Martin-Magniette M-L, Krapp A, Meyer C, Fagard M. Nitrogen Limitation Alters the Response of Specific Genes to Biotic Stress. International Journal of Molecular Sciences. 2018; 19(11):3364. https://doi.org/10.3390/ijms19113364
Chicago/Turabian StyleFarjad, Mahsa, Martine Rigault, Stéphanie Pateyron, Marie-Laure Martin-Magniette, Anne Krapp, Christian Meyer, and Mathilde Fagard. 2018. "Nitrogen Limitation Alters the Response of Specific Genes to Biotic Stress" International Journal of Molecular Sciences 19, no. 11: 3364. https://doi.org/10.3390/ijms19113364
APA StyleFarjad, M., Rigault, M., Pateyron, S., Martin-Magniette, M. -L., Krapp, A., Meyer, C., & Fagard, M. (2018). Nitrogen Limitation Alters the Response of Specific Genes to Biotic Stress. International Journal of Molecular Sciences, 19(11), 3364. https://doi.org/10.3390/ijms19113364