Oncogenic H-Ras Expression Induces Fatty Acid Profile Changes in Human Fibroblasts and Extracellular Vesicles
Abstract
:1. Introduction
2. Results
2.1. Analysis of H-RasV12 and Control Fibroblasts Released EVs by Immunoblotting and Immuno-TEM
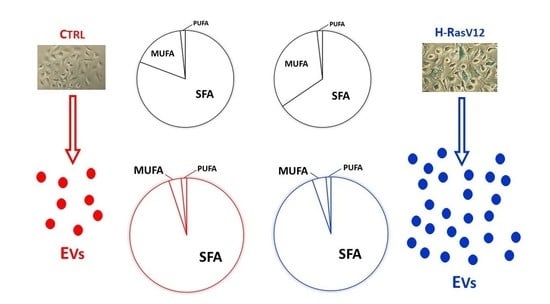
2.2. Analysis of Fatty Acids Content
2.3. Expression Analysis of Genes Involved in Fatty Acids Remodelling and Phospholipases A2
3. Discussion
4. Materials and Methods
4.1. HuDe Fibroblasts Transfection and H-RasV12 Expression
4.2. Staining of Senescence Associated β-Galactosidase (SA-β-Gal)
4.3. Immunostaining
4.4. Extracellular Vesicles Preparation
4.5. Immunoblotting
4.6. Immunogold
4.7. Cells and Vesicles Preparation for Fatty Acid Analysis
4.8. qRT-PCR
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yáñez-Mó, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnstone, R.M.; Adam, M.; Hammond, J.R.; Orr, L.; Turbide, C. Vesicle formation during reticulocyte maturation. Association of plasma membrane activities with released vesicles (exosomes). J. Biol. Chem. 1987, 262, 9412–9420. [Google Scholar] [PubMed]
- Urbanelli, L.; Magini, A.; Buratta, S.; Brozzi, A.; Sagini, K.; Polchi, A.; Tancini, B.; Emiliani, C. Signaling pathways in exosomes biogenesis, secretion and fate. Genes 2013, 4, 152–170. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Lotvall, J.; Hill, A.F.; Hochberg, F.; Buzas, E.I.; Di Vizio, D.; Gardiner, C.; Gho, Y.S.; Kurochkin, I.V.; Mathivanan, S.; Quesenberry, P.; et al. Minimal experimental requirements for definition of extracellular vesicles and their functions: A position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2014, 3, 26913. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Urbanelli, L.; Buratta, S.; Sagini, K.; Ferrara, G.; Lanni, M.; Emiliani, C. Exosome-based strategies for diagnosis and therapy. Recent Pat. CNS Drug Discov. 2015, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.K.; Lee, J.; Kim, S.R.; Choi, D.S.; Yoon, Y.J.; Kim, J.H.; Go, G.; Nhung, D.; Hong, K.; Jang, S.C.; et al. EVpedia: A community web portal for extracellular vesicles research. Bioinformatics 2015, 31, 933–939. [Google Scholar] [CrossRef] [PubMed]
- Laulagnier, K.; Motta, C.; Hamdi, S.; Roy, S.; Fauvelle, F.; Pageaux, J.F.; Kobayashi, T.; Salles, J.P.; Perret, B.; Bonnerot, C.; et al. Mast cell- and dendritic cell-derived exosomes display a specific lipid composition and an unusual membrane organization. Biochem. J. 2004, 380, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Llorente, A.; Skotland, T.; Sylvänne, T.; Kauhanen, D.; Róg, T.; Orłowski, A.; Vattulainen, I.; Ekroos, K.; Sandvig, K. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochim. Biophys. Acta 2013, 1831, 1302–1309. [Google Scholar] [CrossRef] [PubMed]
- Lydic, T.A.; Townsend, S.; Adda, C.G.; Collins, C.; Mathivanan, S.; Reid, G.E. Rapid and comprehensive ‘shotgun’ lipidome profiling of colorectal cancer cell derived exosomes. Methods 2015, 87, 83–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buratta, S.; Urbanelli, L.; Sagini, K.; Giovagnoli, S.; Caponi, S.; Fioretto, D.; Mitro, N.; Caruso, D.; Emiliani, C. Extracellular vesicles released by fibroblasts undergoing H-Ras induced senescence show changes in lipid profile. PLoS ONE 2017, 12, e0188840. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Silvente-Poirot, S.; Poirot, M.; Wakelam, M.J.O. Extracellular vesicles: Lipids as key components of their biogenesis and functions. J. Lipid Res. 2018, 59, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Sagini, K.; Costanzi, E.; Emiliani, C.; Buratta, S.; Urbanelli, L. Extracellular Vesicles as Conveyors of Membrane-Derived Bioactive Lipids in Immune System. Int. J. Mol. Sci. 2018, 19, 1227. [Google Scholar] [CrossRef] [PubMed]
- Boilard, E. Extracellular vesicles and their content in bioactive lipid mediators: More than a sack of microRNA. J. Lipid Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; De Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010, 51, 2105–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esser, J.; Gehrmann, U.; D’Alexandri, F.L.; Hidalgo-Estévez, A.M.; Wheelock, C.E.; Scheynius, A.; Gabrielsson, S.; Rådmark, O. Exosomes from human macrophages and dendritic cells contain enzymes for leukotriene biosynthesis and promote granulocyte migration. J. Allergy Clin. Immunol. 2010, 126, 1032–1040. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.B.; Zhuang, X.; Ju, S.; Xiang, X.; Mu, J.; Liu, Y.; Jiang, H.; Zhang, L.; Mobley, J.; McClain, C.; et al. Exosome-like nanoparticles from intestinal mucosal cells carry prostaglandin E2 and suppress activation of liver NKT cells. J. Immunol. 2013, 190, 3579–3589. [Google Scholar] [CrossRef] [PubMed]
- Record, M.; Carayon, K.; Poirot, M.; Silvente-Poirot, S. Exosomes as new vesicular lipid transporters involved in cell-cell communication and various pathophysiologies. Biochim. Biophys. Acta 2014, 1841, 108–120. [Google Scholar] [CrossRef] [PubMed]
- Lazar, I.; Clement, E.; Attane, C.; Muller, C.; Nieto, L. A new role for extracellular vesicles: How small vesicles can feed tumors’ big appetite. J. Lipid Res. 2018. [Google Scholar] [CrossRef] [PubMed]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Childs, B.G.; Baker, D.J.; Kirkland, J.L.; Campisi, J.; van Deursen, J.M. Senescence and apoptosis: Dueling or complementary cell fates? EMBO Rep. 2014, 15, 1139–1153. [Google Scholar] [CrossRef] [PubMed]
- Lecot, P.; Alimirah, F.; Desprez, P.Y.; Campisi, J.; Wiley, C. Context-dependent effects of cellular senescence in cancer development. Br. J. Cancer 2016, 114, 1180–1184. [Google Scholar] [CrossRef] [PubMed]
- Armeni, T.; Ercolani, L.; Urbanelli, L.; Magini, A.; Magherini, F.; Pugnalono, A.; Piva, F.; Modesti, A.; Emiliani, C.; Principato, G. Cellular redox imbalance and changes of protein S-glutathionylation patterns are associated with senescence induced by oncogenic H-Ras. PLoS ONE 2012, 7, e52151. [Google Scholar] [CrossRef] [PubMed]
- Urbanelli, L.; Magini, A.; Ercolani, L.; Sagini, K.; Polchi, A.; Tancini, B.; Brozzi, A.; Armeni, T.; Principato, G.; Emiliani, C. Oncogenic H-Ras up-regulates acid β-hexosaminidase by a mechanism dependent on the autophagy regulator TFEB. PLoS ONE 2014, 9, e89485. [Google Scholar] [CrossRef] [PubMed]
- Georgilis, A.; Klotz, S.; Hanley, C.J.; Herranz, N.; Weirich, B.; Morancho, B.; Leote, A.C.; D’Artista, L.; Gallage, S.; Seehawer, M.; et al. PTBP1-mediated alternative splicing regulates the inflammatory secretome and the pro-tumorigenic effects of senescent cells. Cancer Cell 2018, 34, 85–102. [Google Scholar] [CrossRef] [PubMed]
- Urbanelli, L.; Buratta, S.; Sagini, K.; Tancini, B.; Emiliani, C. Extracellular Vesicles as New Players in Cellular Senescence. Int. J. Mol. Sci. 2016, 17, 1408. [Google Scholar] [CrossRef] [PubMed]
- Di Micco, R.; Fumagalli, M.; d’Adda di Fagagna, F. Breaking news: High-speed race ends in arrest—How oncogenes induce senescence. Trends Cell Biol. 2007, 17, 529–536. [Google Scholar] [CrossRef] [PubMed]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D.; Lowe, S.W. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Paine, M.S.; Brooks, A.M.; McCubrey, J.A.; Renegar, R.H.; Wang, R.; Terrian, D.M. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008, 68, 7864–7871. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Harris, S.L.; Levine, A.J. The regulation of exosome secretion: A novel function of the p53 protein. Cancer Res. 2006, 66, 4795–4801. [Google Scholar] [CrossRef] [PubMed]
- Osteikoetxea, X.; Balogh, A.; Szabó-Taylor, K.; Németh, A.; Szabó, T.G.; Pálóczi, K.; Sódar, B.; Kittel, Á.; György, B.; Pállinger, É.; et al. Improved Characterization of EV Preparations Based on Protein to Lipid Ratio and Lipid Properties. PLoS ONE 2015, 10, e0121184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marion-Letellier, R.; Savoye, G.; Ghosh, S. Polyunsaturated fatty acids and inflammation. IUBMB Life 2015, 67, 659–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiang, X.; Poliakov, A.; Liu, C.; Liu, Y.; Deng, Z.B.; Wang, J.; Cheng, Z.; Shah, S.V.; Wang, G.J.; Zhang, L.; et al. Induction of myeloid-derived suppressor cells by tumor exosomes. Int. J. Cancer 2009, 124, 2621–2633. [Google Scholar] [CrossRef] [PubMed]
- Kakazu, E.; Mauer, A.S.; Yin, M.; Malhi, H. Hepatocytes release ceramide-enriched pro-inflammatory extracellular vesicles in an IRE1α-dependent manner. J. Lipid Res. 2016, 57, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Hirsova, P.; Ibrahim, S.H.; Krishnan, A.; Verma, V.K.; Bronk, S.F.; Werneburg, N.W.; Charlton, M.R.; Shah, V.H.; Malhi, H.; Gores, G.J. Lipid-induced signaling causes release of inflammatory extracellular vesicles from hepatocytes. Gastroenterology 2016, 150, 956–967. [Google Scholar] [CrossRef] [PubMed]
- Subra, C.; Laulagnier, K.; Perret, B.; Record, M. Exosome lipidomics unravels lipid sorting at the level of multivesicular bodies. Biochimie 2007, 89, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Yang, X.F.; Liu, H.L.; Fu, N.; Ouyang, Y.; Qing, K. Long-chain acyl-CoA synthetase in fatty acid metabolism involved in liver and other diseases: An update. World J. Gastroenterol. 2015, 21, 3492–3498. [Google Scholar] [CrossRef] [PubMed]
- Golej, D.L.; Askari, B.; Kramer, F.; Barnhart, S.; Vivekanandan-Giri, A.; Pennathur, S.; Bornfeldt, K.E. Long-chain acyl-CoA synthetase 4 modulates prostaglandin E2 release from human arterial smooth muscle cells. J. Lipid Res. 2011, 52, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Radif, Y.; Ndiaye, H.; Kalantzi, V.; Jacobs, R.; Hall, A.; Minogue, S.; Waugh, M.G. The endogenous subcellular localisations of the long chain fatty acid-activating enzymes ACSL3 and ACSL4 in sarcoma and breast cancer cells. Mol. Cell. Biochem. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cadenas, C.; Vosbeck, S.; Hein, E.M.; Hellwig, B.; Langer, A.; Hayen, H.; Franckenstein, D.; Büttner, B.; Hammad, S.; Marchan, R.; et al. Glycerophospholipid profile in oncogene-induced senescence. Biochim. Biophys. Acta 2012, 1821, 1256–1268. [Google Scholar] [CrossRef] [PubMed]
- Lizardo, D.Y.; Lin, Y.L.; Gokcumen, O.; Atilla-Gokcumen, G.E. Regulation of lipids is central to replicative senescence. Mol. Biosyst. 2017, 13, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 enzymes: Physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Sato, H.; Miki, Y.; Yamamoto, K.; Taketomi, Y. A new era of secreted phospholipase A2. J. Lipid Res. 2015, 56, 1248–1261. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Masuda, S.; Shimbara, S.; Ishikawa, Y.; Ishii, T.; Kudo, Y. Cellular distribution, post-translational modification, and tumorigenic potential of human group III secreted phospholipase A2. J. Biol. Chem. 2005, 280, 24987–24998. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Masuda, S.; Ueda-Semmyo, K.; Yoda, E.; Kuwata, H.; Takanezawa, Y.; Aoki, J.; Arai, H.; Sumimoto, H.; Ishikawa, Y.; et al. Group VIB Ca2+-independent phospholipase A2γ promotes cellular membrane hydrolysis and prostaglandin production in a manner distinct from other intracellular phospholipases A2. J. Biol. Chem. 2005, 280, 14028–14041. [Google Scholar] [CrossRef] [PubMed]
- Cermenati, G.; Giatti, S.; Audano, M.; Pesaresi, M.; Spezzano, R.; Caruso, D.; Mitro, N.; Melcangi, R.C. Diabetes alters myelin lipid profile in rat cerebral cortex: Protective effects of dihydroprogesterone. J. Steroid Biochem. Mol. Biol. 2017, 168, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Cermenati, G.; Audano, M.; Giatti, S.; Carozzi, V.; Porretta-Serapiglia, C.; Pettinato, E.; Ferri, C.; D’Antonio, M.; De Fabiani, E.; Crestani, M.; et al. Lack of sterol regulatory element binding factor-1c imposes glial Fatty Acid utilization leading to peripheral neuropathy. Cell Metab. 2015, 21, 571–583. [Google Scholar] [CrossRef] [PubMed]





| ng Lipid/µg Protein | Fibroblasts | EVs | ||
|---|---|---|---|---|
| CTRL | RasV12 | CTRL | RasV12 | |
| SFA | 212.39 ± 66.30 | 126.19 ± 47.05 * | 1084.78 ± 147.63 | 787.4 ± 65.65 * |
| MUFA | 35.14 ± 12.29 | 70.60 ± 31.65 * | 40.38 ± 8.19 | 32.28 ± 7.22 |
| PUFA | 3.35 ± 1.54 | 3.38 ± 1.48 | 18.35 ± 8.46 | 11.67 ± 4.19 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sagini, K.; Urbanelli, L.; Costanzi, E.; Mitro, N.; Caruso, D.; Emiliani, C.; Buratta, S. Oncogenic H-Ras Expression Induces Fatty Acid Profile Changes in Human Fibroblasts and Extracellular Vesicles. Int. J. Mol. Sci. 2018, 19, 3515. https://doi.org/10.3390/ijms19113515
Sagini K, Urbanelli L, Costanzi E, Mitro N, Caruso D, Emiliani C, Buratta S. Oncogenic H-Ras Expression Induces Fatty Acid Profile Changes in Human Fibroblasts and Extracellular Vesicles. International Journal of Molecular Sciences. 2018; 19(11):3515. https://doi.org/10.3390/ijms19113515
Chicago/Turabian StyleSagini, Krizia, Lorena Urbanelli, Eva Costanzi, Nico Mitro, Donatella Caruso, Carla Emiliani, and Sandra Buratta. 2018. "Oncogenic H-Ras Expression Induces Fatty Acid Profile Changes in Human Fibroblasts and Extracellular Vesicles" International Journal of Molecular Sciences 19, no. 11: 3515. https://doi.org/10.3390/ijms19113515
APA StyleSagini, K., Urbanelli, L., Costanzi, E., Mitro, N., Caruso, D., Emiliani, C., & Buratta, S. (2018). Oncogenic H-Ras Expression Induces Fatty Acid Profile Changes in Human Fibroblasts and Extracellular Vesicles. International Journal of Molecular Sciences, 19(11), 3515. https://doi.org/10.3390/ijms19113515