Genome-Wide Transcriptional and Functional Analysis of Human T Lymphocytes Treated with Benzo[α]pyrene
Abstract
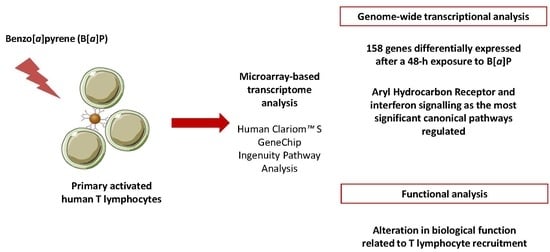
:1. Introduction
2. Results
2.1. B[α]P Exposure Alters the Expression of 158 Genes in Human T Lymphocytes
2.2. The AhR and IFN Signalling are the Most Significant Canonical Pathways Regulated by B[α]P in Human T Lymphocytes
2.3. IPA Functional Analysis Revealed the Prominence of Categories Related to Cellular Movement for B[α]P-Regulated Genes in Human T Lymphocytes
2.4. B[α]P Inhibits Human T Lymphocyte Chemotaxis and Transendothelial Migration
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Microarray Experiments
4.2.1. RNA Extraction
4.2.2. Microarray Hybridization
4.2.3. Data Normalization
4.3. Statistical Filtration of Differentially Expressed Genes
4.4. Functional Analysis by Ingenuity Pathway Analysis (IPA)
4.5. RT-qPCR Assays
4.6. Chemotaxis and Transendothelial Migration Assays
4.7. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Hattemer-Frey, H.A.; Travis, C.C. Benzo-a-pyrene: Environmental partitioning and human exposure. Toxicol. Ind. Health 1991, 7, 141–157. [Google Scholar] [CrossRef] [PubMed]
- Clark, J.D.; Serdar, B.; Lee, D.J.; Arheart, K.; Wilkinson, J.D.; Fleming, L.E. Exposure to polycyclic aromatic hydrocarbons and serum inflammatory markers of cardiovascular disease. Environ. Res. 2012, 117, 132–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, H. Synergistic mechanisms in carcinogenesis by polycyclic aromatic hydrocarbons and by tobacco smoke: A bio-historical perspective with updates. Carcinogenesis 2001, 22, 1903–1930. [Google Scholar] [CrossRef] [PubMed]
- Hankinson, O. The aryl hydrocarbon receptor complex. Annu. Rev. Pharmacol. Toxicol. 1995, 35, 307–340. [Google Scholar] [CrossRef] [PubMed]
- De Vries, A.; Dollé, M.E.; Broekhof, J.L.; Muller, J.J.; Kroese, E.D.; van Kreijl, C.F.; Capel, P.J.; Vijg, J.; van Steeg, H. Induction of DNA adducts and mutations in spleen, liver and lung of XPA-deficient/lacZ transgenic mice after oral treatment with benzo[a]pyrene: Correlation with tumour development. Carcinogenesis 1997, 18, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Okano, P.; Miller, H.N.; Robinson, R.C.; Gelboin, H.V. Comparison of benzo(a)pyrene and (-)-trans-7,8-dihydroxy-7,8-dihydrobenzo(a)pyrene metabolism in human blood monocytes and lymphocytes. Cancer Res. 1979, 39, 3184–3193. [Google Scholar] [PubMed]
- Prigent, L.; Robineau, M.; Jouneau, S.; Morzadec, C.; Louarn, L.; Vernhet, L.; Fardel, O.; Sparfel, L. The aryl hydrocarbon receptor is functionally upregulated early in the course of human T-cell activation. Eur. J. Immunol. 2014, 44, 1330–1340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, K.L.; Lysy, H.H.; Holsapple, M.P. Immunosuppression by polycyclic aromatic hydrocarbons: A structure-activity relationship in B6C3F1 and DBA/2 mice. Immunopharmacology 1985, 9, 155–164. [Google Scholar] [CrossRef]
- Wojdani, A.; Alfred, L.J. Alterations in cell-mediated immune functions induced in mouse splenic lymphocytes by polycyclic aromatic hydrocarbons. Cancer Res. 1984, 44, 942–945. [Google Scholar] [PubMed]
- Selgrade, M.J.; Daniels, M.J.; Burleson, G.R.; Lauer, L.D.; Dean, J.H. Effects of 7,12-dimethylbenz[a]anthracene, benzo[a]pyrene and cyclosporin A on murine cytomegalovirus infection: Studies of resistance mechanisms. Int. J. Immunopharmacol. 1988, 10, 811–818. [Google Scholar] [CrossRef]
- Dean, J.H.; Luster, M.I.; Boorman, G.A.; Lauer, L.D.; Ward, E.C. Immunotoxicity of Tumor Promoting Environmental Chemicals and Phorbol Diesters. In Advances in Immunopharmacology; Hadden, J.W., Chedid, L., Dukor, P., Spreafico, F., Willoughby, D., Eds.; Pergamon: Amsterdam, The Netherlands, 1983; ISBN 978-0-08-029775-0. [Google Scholar]
- Davila, D.R.; Romero, D.L.; Burchiel, S.W. Human T cells are highly sensitive to suppression of mitogenesis by polycyclic aromatic hydrocarbons and this effect is differentially reversed by α-naphthoflavone. Toxicol. Appl. Pharmacol. 1996, 139, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Mudzinski, S.P. Effects of benzo[a]pyrene on concanavalin A-stimulated human peripheral blood mononuclear cells in vitro: Inhibition of proliferation but no effect on parameters related to the G1 phase of the cell cycle. Toxicol. Appl. Pharmacol. 1993, 119, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Gammon, M.D.; Santella, R.M.; Neugut, A.I.; Eng, S.M.; Teitelbaum, S.L.; Paykin, A.; Levin, B.; Terry, M.B.; Young, T.L.; Wang, L.W.; et al. Environmental toxins and breast cancer on Long Island. I. Polycyclic aromatic hydrocarbon DNA adducts. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 2002, 11, 677–685. [Google Scholar]
- Li, D.; Wang, M.; Cheng, L.; Spitz, M.R.; Hittelman, W.N.; Wei, Q. In vitro induction of benzo(a)pyrene diol epoxide-DNA adducts in peripheral lymphocytes as a susceptibility marker for human lung cancer. Cancer Res. 1996, 56, 3638–3641. [Google Scholar] [PubMed]
- Liamin, M.; Boutet-Robinet, E.; Jamin, E.L.; Fernier, M.; Khoury, L.; Kopp, B.; Le Ferrec, E.; Vignard, J.; Audebert, M.; Sparfel, L. Benzo[α]pyrene-induced DNA damage associated with mutagenesis in primary human activated T lymphocytes. Biochem. Pharmacol. 2017, 137, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Hockley, S.L.; Arlt, V.M.; Brewer, D.; Giddings, I.; Phillips, D.H. Time- and concentration-dependent changes in gene expression induced by benzo(α)pyrene in two human cell lines, MCF-7 and HepG2. BMC Genom. 2006, 7, 260. [Google Scholar] [CrossRef] [PubMed]
- Sparfel, L.; Pinel-Marie, M.-L.; Boize, M.; Koscielny, S.; Desmots, S.; Pery, A.; Fardel, O. Transcriptional signature of human macrophages exposed to the environmental contaminant benzo(α)pyrene. Toxicol. Sci. Off. J. Soc. Toxicol. 2010, 114, 247–259. [Google Scholar] [CrossRef] [PubMed]
- Bernshausen, T.; Jux, B.; Esser, C.; Abel, J.; Fritsche, E. Tissue distribution and function of the Aryl hydrocarbon receptor repressor (AhRR) in C57BL/6 and Aryl hydrocarbon receptor deficient mice. Arch. Toxicol. 2006, 80, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Brauze, D.; Zawierucha, P.; Kiwerska, K.; Bednarek, K.; Oleszak, M.; Rydzanicz, M.; Jarmuz-Szymczak, M. Induction of expression of aryl hydrocarbon receptor-dependent genes in human HepaRG cell line modified by shRNA and treated with β-naphthoflavone. Mol. Cell Biochem. 2017, 425, 59–75. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Wang, A.; Mauro, C.; Marelli-Berg, F. T lymphocyte trafficking: Molecules and mechanisms. Front. Biosci. Landmark Ed. 2013, 18, 422–440. [Google Scholar] [PubMed]
- Munk, R.; Ghosh, P.; Ghosh, M.C.; Saito, T.; Xu, M.; Carter, A.; Indig, F.; Taub, D.D.; Longo, D.L. Involvement of mTOR in CXCL12 mediated T cell signaling and migration. PLoS ONE 2011, 6, e24667. [Google Scholar] [CrossRef] [PubMed]
- Ticchioni, M.; Charvet, C.; Noraz, N.; Lamy, L.; Steinberg, M.; Bernard, A.; Deckert, M. Signaling through ZAP-70 is required for CXCL12-mediated T-cell transendothelial migration. Blood 2002, 99, 3111–3118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DellaValle, C.T.; Deziel, N.C.; Jones, R.R.; Colt, J.S.; De Roos, A.J.; Cerhan, J.R.; Cozen, W.; Severson, R.K.; Flory, A.R.; Morton, L.M.; et al. Polycyclic aromatic hydrocarbons: determinants of residential carpet dust levels and risk of non-Hodgkin lymphoma. Cancer Causes Control CCC 2016, 27, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Raghavan, A.; Ogilvie, R.L.; Reilly, C.; Abelson, M.L.; Raghavan, S.; Vasdewani, J.; Krathwohl, M.; Bohjanen, P.R. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002, 30, 5529–5538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambroise, J.; Bearzatto, B.; Robert, A.; Govaerts, B.; Macq, B.; Gala, J.-L. Impact of the spotted microarray preprocessing method on fold-change compression and variance stability. BMC Bioinform. 2011, 12, 413. [Google Scholar] [CrossRef] [PubMed]
- Hockley, S.L.; Arlt, V.M.; Brewer, D.; Te Poele, R.; Workman, P.; Giddings, I.; Phillips, D.H. AHR- and DNA-damage-mediated gene expression responses induced by benzo(a)pyrene in human cell lines. Chem. Res. Toxicol. 2007, 20, 1797–1810. [Google Scholar] [CrossRef] [PubMed]
- Keshava, C.; Whipkey, D.; Weston, A. Transcriptional signatures of environmentally relevant exposures in normal human mammary epithelial cells: benzo[a]pyrene. Cancer Lett. 2005, 221, 201–211. [Google Scholar] [CrossRef] [PubMed]
- Keshava, C.; Divi, R.L.; Einem, T.L.; Richardson, D.L.; Leonard, S.L.; Keshava, N.; Poirier, M.C.; Weston, A. Chlorophyllin significantly reduces benzo[a]pyrene-DNA adduct formation and alters cytochrome P450 1A1 and 1B1 expression and EROD activity in normal human mammary epithelial cells. Environ. Mol. Mutagen. 2009, 50, 134–144. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Shao, J.; Li, H.; Yu, Y. Early whole-genome transcriptional response induced by benzo[a]pyrene diol epoxide in a normal human cell line. Genomics 2009, 93, 332–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frericks, M.; Meissner, M.; Esser, C. Microarray analysis of the AHR system: tissue-specific flexibility in signal and target genes. Toxicol. Appl. Pharmacol. 2007, 220, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Brinchmann, B.C.; Le Ferrec, E.; Podechard, N.; Lagadic-Gossmann, D.; Shoji, K.F.; Penna, A.; Kukowski, K.; Kubátová, A.; Holme, J.A.; Øvrevik, J. Lipophilic Chemicals from Diesel Exhaust Particles Trigger Calcium Response in Human Endothelial Cells via Aryl Hydrocarbon Receptor Non-Genomic Signalling. Int. J. Mol. Sci. 2018, 19, 1429. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Takaoka, A.; Taniguchi, T. Type I interferon [corrected] gene induction by the interferon regulatory factor family of transcription factors. Immunity 2006, 25, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Horimoto, H.; Kameyama, T.; Hayakawa, S.; Yamato, H.; Dazai, M.; Takada, A.; Kida, H.; Bott, D.; Zhou, A.C.; et al. Constitutive aryl hydrocarbon receptor signaling constrains type I interferon–mediated antiviral innate defense. Nat. Immunol. 2016, 17, 687–694. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, S.; Huang, Y.; Feng, Z.; Xu, L.; Jin, Y.; Lu, J. The toxic effects of benzo[a]pyrene on activated mouse T cells in vitro. Immunopharmacol. Immunotoxicol. 2017, 39, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Ciencewicki, J.; Jaspers, I. Air Pollution and Respiratory Viral Infection. Inhal. Toxicol. 2007, 19, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.; Im, H.; Oh, E.; Lee, J.; Khim, J.; Mun, J.; Kim, Y.; Lee, E.; Kim, J.; Sul, D. Effects of benzo(a)pyrene on protein expression in Jurkat T-cells. Proteomics 2004, 4, 3514–3526. [Google Scholar] [CrossRef] [PubMed]
- Trifilo, M.J.; Bergmann, C.C.; Kuziel, W.A.; Lane, T.E. CC chemokine ligand 3 (CCL3) regulates CD8(+)-T-cell effector function and migration following viral infection. J. Virol. 2003, 77, 4004–4014. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Pan, J.; Setiadi, H.; Patel, K.D.; McEver, R.P. Interleukin 4 or oncostatin M induces a prolonged increase in P-selectin mRNA and protein in human endothelial cells. J. Exp. Med. 1996, 184, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Nedelkovska, H.; Rosenberg, A.F.; Hilchey, S.P.; Hyrien, O.; Burack, W.R.; Quataert, S.A.; Baker, C.M.; Azadniv, M.; Welle, S.L.; Ansell, S.M.; et al. Follicular Lymphoma Tregs Have a Distinct Transcription Profile Impacting Their Migration and Retention in the Malignant Lymph Node. PLoS ONE 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Bae, Y.S.; Park, J.C.; Suh, P.G.; Ryu, S.H. The synthetic peptide, His-Phe-Tyr-Leu-Pro-Met, is a chemoattractant for Jukat T cells. Exp. Mol. Med. 2001, 33, 257–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, P.M.; Park, M.S.; Chow, M.; Chang, J.H.; Wrischnik, L.; Chan, W.K. Benzo[a]pyrene increases the Nrf2 content by downregulating the Keap1 message. Toxicol. Sci. Off. J. Soc. Toxicol. 2010, 116, 549–561. [Google Scholar] [CrossRef] [PubMed]
- Amiot, L.; Vu, N.; Rauch, M.; L’Helgoualc’h, A.; Chalmel, F.; Gascan, H.; Turlin, B.; Guyader, D.; Samson, M. Expression of HLA-G by mast cells is associated with hepatitis C virus-induced liver fibrosis. J. Hepatol. 2014, 60, 245–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, G.R.; Masri, S.H.; David, R.; Jones, M.; Datta, A.; Lombardi, G.; Runkell, L.; de Dios, C.; Sizing, I.; James, M.J.; et al. IFN-α Subtypes Differentially Affect Human T Cell Motility. J. Immunol. 2004, 173, 1663–1670. [Google Scholar] [CrossRef] [PubMed]
- De Waard, W.J.; Aarts, J.M.M.J.G.; Peijnenburg, A.C.M.; Baykus, H.; Talsma, E.; Punt, A.; de Kok, T.M.C.M.; van Schooten, F.J.; Hoogenboom, L.P. Gene expression profiling in Caco-2 human colon cells exposed to TCDD, benzo[a]pyrene, and natural Ah receptor agonists from cruciferous vegetables and citrus fruits. Toxicol. In Vitro Int. J. Publ. Assoc. BIBRA 2008, 22, 396–410. [Google Scholar] [CrossRef] [PubMed]
- Kendziorski, C.M.; Zhang, Y.; Lan, H.; Attie, A.D. The efficiency of pooling mRNA in microarray experiments. Biostat. Oxf. Engl. 2003, 4, 465–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piccolo, S.R.; Withers, M.R.; Francis, O.E.; Bild, A.H.; Johnson, W.E. Multiplatform single-sample estimates of transcriptional activation. Proc. Natl. Acad. Sci. USA 2013, 110, 17778–17783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, M.; Wang, P.; Boyd, A.D.; Kostov, G.; Athey, B.; Jones, E.G.; Bunney, W.E.; Myers, R.M.; Speed, T.P.; Akil, H.; et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005, 33, e175. [Google Scholar] [CrossRef] [PubMed]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2013, 41, D991–D995. [Google Scholar] [CrossRef] [PubMed]
- Darde, T.A.; Gaudriault, P.; Beranger, R.; Lancien, C.; Caillarec-Joly, A.; Sallou, O.; Bonvallot, N.; Chevrier, C.; Mazaud-Guittot, S.; Jégou, B.; et al. TOXsIgN: A cross-species repository for toxicogenomic signatures. Bioinform. Oxf. Engl. 2018, 34, 2116–2122. [Google Scholar] [CrossRef] [PubMed]
- Chalmel, F.; Primig, M. The Annotation, Mapping, Expression and Network (AMEN) suite of tools for molecular systems biology. BMC Bioinform. 2008, 9, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayati, A.; Podechard, N.; Rineau, M.; Sparfel, L.; Lagadic-Gossmann, D.; Fardel, O.; Le Ferrec, E. Benzo(a)pyrene triggers desensitization of β2-adrenergic pathway. Sci. Rep. 2017, 7, 3262. [Google Scholar] [CrossRef] [PubMed]





| ID | Gene Name | Description | Differential Expression (log-2) a | p-Value |
|---|---|---|---|---|
| Top 15 up-regulated genes | ||||
| 1543 | CYP1A1 | cytochrome P450, family 1, subfamily A, polypeptide 1 | 1.152 | 5.9 × 10−3 |
| 1545 | CYP1B1 | cytochrome P450, family 1, subfamily B, polypeptide 1 | 1.058 | 4.7 × 10−3 |
| 2706 | GJB2 | gap junction protein, beta 2, 26 kDa | 0.702 | 5.7 × 10−3 |
| 10804 | GJB6 | gap junction protein, beta 6, 30 kDa | 0.528 | 7.7 × 10−3 |
| 25976 | TIPARP | TCDD-inducible poly (ADP-ribose) polymerase | 0.528 | 1.0 × 10−2 |
| 153339 | TMEM167A | transmembrane protein 167A | 0.491 | 3.0 × 10−2 |
| 167826 | OLIG3 | oligodendrocyte transcription factor 3 | 0.484 | 2.7 × 10−2 |
| 91768 | CABLES1 | Cdk5 and Abl enzyme substrate 1 | 0.426 | 3.1 × 10−4 |
| 3815 | KIT | proto-oncogene c-Kit | 0.401 | 1.2 × 10−5 |
| 51676 | ASB2 | ankyrin repeat and SOCS box containing 2 | 0.388 | 5.6 × 10−4 |
| 83888 | FGFBP2 | fibroblast growth factor binding protein 2 | 0.362 | 2.0 × 10-3 |
| 5774 | PTPN3 | protein tyrosine phosphatase, non-receptor type 3 | 0.349 | 1.1 × 10−2 |
| 9289 | ADGRG1 | adhesion G protein-coupled receptor G1 | 0.346 | 5.8 × 10-3 |
| 23682 | RAB38 | RAB38, member RAS oncogene family | 0.328 | 4.6 × 10−3 |
| 81618 | ITM2C | integral membrane protein 2C | 0.307 | 1.5 × 10−2 |
| Top 15 down-regulated genes | ||||
| 10964 | IFI44L | interferon-induced protein 44-like | −0.451 | 1.7 × 10−2 |
| 1293 | COL6A3 | collagen, type VI, alpha 3 | −0.337 | 6.3 × 10−3 |
| 10561 | IFI44 | interferon-induced protein 44 | −0.322 | 3.2 × 10−2 |
| 79648 | MCPH1 | microcephalin 1 | −0.319 | 1.0 × 10−2 |
| 1130 | LYST | lysosomal trafficking regulator | −0.282 | 2.2 × 10−2 |
| 4599 | MX1 | MX dynamin-like GTPase 1 | −0.282 | 4.8 × 10−2 |
| 8638 | OASL | 2′-5′-oligoadenylate synthetase- | −0.269 | 2.0 × 10−3 |
| 3433 | IFIT2 | interferon-induced protein with tetratricopeptide repeats 2 | −0.267 | 5.6 × 10−4 |
| 10216 | PRG4 | proteoglycan 4 | −0.266 | 3.4 × 10−2 |
| 7070 | THY1 | Thy-1 cell surface antigen | −0.265 | 1.0 × 10−2 |
| 6402 | SELL | selectin L | −0.233 | 3.1 × 10−2 |
| 56479 | KCNQ5 | potassium channel, voltage gated KQT-like subfamily Q, member 5 | −0.232 | 1.7 × 10−2 |
| 1803 | DPP4 | dipeptidyl-peptidase 4 | −0.222 | 2.3 × 10−2 |
| 3394 | IRF8 | interferon regulatory factor 8 | −0.220 | 1.2 × 10−2 |
| 5167 | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | −0.207 | 2.1 × 10−2 |
| Top Pathways | ap-Value | b Regulated Genes |
|---|---|---|
| Up-regulated genes | ||
| Aryl Hydrocarbon Receptor Signaling | 3.99 × 10−5 | AhRR, CCNE2, CDKN1A, CYP1A1, CYP1B1, NQO1 |
| Protein Kinase A Signaling | 7.36 × 10−5 | ADCY9, ADD2, DUSP4, MYH10, MYL9, PPP1R14C, PTPDC1, PTPN3, SAMD3 |
| Estrogen-mediated S-phase entry | 2.06 × 10−4 | CCNE2, CDKN1A, E2F7 |
| Cell cycle: G1/S checkpoint regulation | 2.14 × 10−4 | CCNE2, CDKN1A, E2F7, SAMD3 |
| Down-regulated genes | ||
| Interferon Signaling | 1.41 × 10−4 | IFIT3, MX1, OAS1 |
| Granulocyte Adhesion and Diapedesis | 1.74 × 10−4 | CCL3, CCL3L3, CCL4L1, CCL4L2, SELL, THY1 |
| Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses | 5.97 × 10−4 | DDX58, IFIH1, OAS1, OSM |
| Activation of IRF by Cytosolic Pattern Recognition Receptors | 7.42 × 10−4 | DDX58, IFIH1, IFIT2 |
| Network | Top Functions | p-Value a | Focus Genes b |
|---|---|---|---|
| Diseases and Disorders | |||
| 1 | Cancer | 1.82 × 10−3–1.61 × 10−12 | 144 |
| 2 | Hematological Disease | 1.64 × 10−3–1.61 × 10−12 | 67 |
| 3 | Immunological Disease | 1.64 × 10−3–1.61 × 10−12 | 73 |
| 4 | Organismal Injury and Abnormalities | 1.82 × 10−3–1.61 × 10−12 | 147 |
| 5 | Antimicrobial Response | 2.89 × 10−4–2.23 × 10−10 | 19 |
| Molecular and Cellular functions | |||
| 1 | Cellular Movement | 1.77 × 10−3–6.81 × 10−12 | 60 |
| 2 | Cell Death and Survival | 1.59 × 10−3–2.05 × 10−11 | 77 |
| 3 | Cellular Function and Maintenance | 1.30 × 10−3–7.97 × 10−11 | 52 |
| 4 | Cell-To-Cell Signaling and Interaction | 1.82 × 10−3–4.36 × 10−8 | 41 |
| 5 | Cellular Growth and Proliferation | 1.68 × 10−3–5.20 × 10−8 | 77 |
| Physiological system development and function | |||
| 1 | Immune Cell Trafficking | 1.82 × 10−3–6.81 × 10−12 | 45 |
| 2 | Hematological System Development and Function | 1.82 × 10−3–5.20 × 10−10 | 64 |
| 3 | Tissue Morphology | 1.67 × 10−3–2.55 × 10−9 | 56 |
| 4 | Digestive System Development and Function | 1.07 × 10−3–1.75 × 10−7 | 25 |
| 5 | Lymphoid Tissue Structure and Development | 1.78 × 10−3–1.75 × 10−7 | 48 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liamin, M.; Le Mentec, H.; Evrard, B.; Huc, L.; Chalmel, F.; Boutet-Robinet, E.; Le Ferrec, E.; Sparfel, L. Genome-Wide Transcriptional and Functional Analysis of Human T Lymphocytes Treated with Benzo[α]pyrene. Int. J. Mol. Sci. 2018, 19, 3626. https://doi.org/10.3390/ijms19113626
Liamin M, Le Mentec H, Evrard B, Huc L, Chalmel F, Boutet-Robinet E, Le Ferrec E, Sparfel L. Genome-Wide Transcriptional and Functional Analysis of Human T Lymphocytes Treated with Benzo[α]pyrene. International Journal of Molecular Sciences. 2018; 19(11):3626. https://doi.org/10.3390/ijms19113626
Chicago/Turabian StyleLiamin, Marie, Hélène Le Mentec, Bertrand Evrard, Laurence Huc, Frédéric Chalmel, Elisa Boutet-Robinet, Eric Le Ferrec, and Lydie Sparfel. 2018. "Genome-Wide Transcriptional and Functional Analysis of Human T Lymphocytes Treated with Benzo[α]pyrene" International Journal of Molecular Sciences 19, no. 11: 3626. https://doi.org/10.3390/ijms19113626
APA StyleLiamin, M., Le Mentec, H., Evrard, B., Huc, L., Chalmel, F., Boutet-Robinet, E., Le Ferrec, E., & Sparfel, L. (2018). Genome-Wide Transcriptional and Functional Analysis of Human T Lymphocytes Treated with Benzo[α]pyrene. International Journal of Molecular Sciences, 19(11), 3626. https://doi.org/10.3390/ijms19113626