Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions
Abstract
:1. Introduction
2. Results
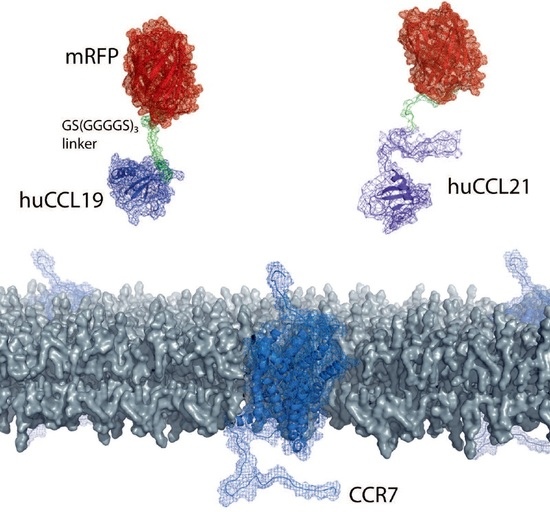
2.1. Design of Fluorescently Tagged CCL19 and CCL21
2.2. Purification and Functional Characterization of CCL19-mRFP and CCL21-mRFP
2.3. CCL19-mRFP Is Internalized by CCR7 and ACKR4, whereas CCL21-mRFP Is Preferentially Scavenged by ACKR4
2.4. Monitoring DC Migration in 3D Collagen along Gradients of CCL19-mRFP or CCL21-mRFP by Time-Lapse Video Microscopy
2.5. Monitoring CCR7-Dependent Cancer Cell Migration in 3D Collagen along a CCL19-mRFP Gradient
3. Discussion
4. Materials and Methods
4.1. Reagents and Antibodies
4.2. Cloning, Expression, and Purification of CCL19-mRFP and CCL21-mRFP
4.3. Cloning and Expression of Chemokine Receptors
4.4. Preparation of Monocyte-Derived and Bone-Marrow-Derived DCs
4.5. Chemotaxis Assays
4.6. Calcium Flux
4.7. β-Arrestin Recruitment Determined by Bioluminescence Resonance Energy Transfer (BRET)
4.8. Confocal Live-Cell Imaging
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ACKR4 | Atypical chemokine receptor 4 |
| CCL19 | CC chemokine ligand 19 |
| CCL21 | CC chemokine ligand 21 |
| CCR7 | CC chemokine receptor 7 |
| DCs | Dendritic cells |
| ERK | Extracellular signaling regulated kinase |
| GAGs | Glycosaminoglycans |
| GPCR | G-protein coupled receptors |
| MoDCs | Monocyte-derived DCs |
| mRFP | Monomeric red fluorescent protein |
| PTx | Pertussis toxin |
References
- Baggiolini, M. Chemokines and leukocyte traffic. Nature 1998, 392, 565–568. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.W.; Sokol, C.L.; Luster, A.D. Chemokines and chemokine receptors: Positioning cells for host defense and immunity. Annu. Rev. Immunol. 2014, 32, 659–702. [Google Scholar] [CrossRef] [PubMed]
- Balkwill, F.R. The chemokine system and cancer. J. Pathol. 2012, 226, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Legler, D.F.; Thelen, M. Chemokines: Chemistry, Biochemistry and Biological Function. Chimia 2016, 70, 856–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Legler, D.F.; Thelen, M. New insights in chemokine signaling. F1000Research 2018, 7, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hauser, M.A.; Schaeuble, K.; Kindinger, I.; Impellizzieri, D.; Krueger, W.A.; Hauck, C.R.; Boyman, O.; Legler, D.F. Inflammation-Induced CCR7 Oligomers Form Scaffolds to Integrate Distinct Signaling Pathways for Efficient Cell Migration. Immunity 2016, 44, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Link, A.; Vogt, T.K.; Favre, S.; Britschgi, M.R.; Acha-Orbea, H.; Hinz, B.; Cyster, J.G.; Luther, S.A. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat. Immunol. 2007, 8, 1255–1265. [Google Scholar] [CrossRef]
- Forster, R.; Davalos-Misslitz, A.C.; Rot, A. CCR7 and its ligands: Balancing immunity and tolerance. Nat. Rev. Immunol. 2008, 8, 362–371. [Google Scholar] [CrossRef]
- Hauser, M.A.; Legler, D.F. Common and biased signaling pathways of the chemokine receptor CCR7 elicited by its ligands CCL19 and CCL21 in leukocytes. J. Leukocyte Biol. 2016, 99, 869–882. [Google Scholar] [CrossRef] [Green Version]
- Moschovakis, G.L.; Forster, R. Multifaceted activities of CCR7 regulate T-cell homeostasis in health and disease. Eur. J. Immunol. 2012, 42, 1949–1955. [Google Scholar] [CrossRef] [Green Version]
- Legler, D.F.; Uetz-von Allmen, E.; Hauser, M.A. CCR7: Roles in cancer cell dissemination, migration and metastasis formation. Int. J. Biochem. Cell Biol. 2014, 54C, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Otero, C.; Groettrup, M.; Legler, D.F. Opposite Fate of Endocytosed CCR7 and Its Ligands: Recycling versus Degradation. J. Immunol. 2006, 177, 2314–2323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schumann, K.; Lammermann, T.; Bruckner, M.; Legler, D.F.; Polleux, J.; Spatz, J.P.; Schuler, G.; Forster, R.; Lutz, M.B.; Sorokin, L.; et al. Immobilized chemokine fields and soluble chemokine gradients cooperatively shape migration patterns of dendritic cells. Immunity 2010, 32, 703–713. [Google Scholar] [CrossRef] [PubMed]
- Laufer, J.M.; Legler, D.F. Beyond migration-Chemokines in lymphocyte priming, differentiation, and modulating effector functions. J. Leukocyte Biol. 2018, 104, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Weber, M.; Hauschild, R.; Schwarz, J.; Moussion, C.; de Vries, I.; Legler, D.F.; Luther, S.A.; Bollenbach, T.; Sixt, M. Interstitial dendritic cell guidance by haptotactic chemokine gradients. Science 2013, 339, 328–332. [Google Scholar] [CrossRef] [PubMed]
- Russo, E.; Teijeira, A.; Vaahtomeri, K.; Willrodt, A.H.; Bloch, J.S.; Nitschke, M.; Santambrogio, L.; Kerjaschki, D.; Sixt, M.; Halin, C. Intralymphatic CCL21 Promotes Tissue Egress of Dendritic Cells through Afferent Lymphatic Vessels. Cell Rep. 2016, 14, 1723–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaahtomeri, K.; Brown, M.; Hauschild, R.; de Vries, I.; Leithner, A.F.; Mehling, M.; Kaufmann, W.A.; Sixt, M. Locally Triggered Release of the Chemokine CCL21 Promotes Dendritic Cell Transmigration across Lymphatic Endothelia. Cell Rep. 2017, 19, 902–909. [Google Scholar] [CrossRef] [PubMed]
- Hirose, J.; Kawashima, H.; Swope Willis, M.; Springer, T.A.; Hasegawa, H.; Yoshie, O.; Miyasaka, M. Chondroitin sulfate B exerts its inhibitory effect on secondary lymphoid tissue chemokine (SLC) by binding to the C-terminus of SLC. Biochim. Biophys. Acta 2002, 1571, 219–224. [Google Scholar] [CrossRef]
- Hauser, M.A.; Kindinger, I.; Laufer, J.M.; Spate, A.K.; Bucher, D.; Vanes, S.L.; Krueger, W.A.; Wittmann, V.; Legler, D.F. Distinct CCR7 glycosylation pattern shapes receptor signaling and endocytosis to modulate chemotactic responses. J. Leukocyte Biol. 2016, 99, 993–1007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proudfoot, A.E.I.; Johnson, Z.; Bonvin, P.; Handel, T.M. Glycosaminoglycan Interactions with Chemokines Add Complexity to a Complex System. Pharmaceuticals 2017, 10, E70. [Google Scholar] [CrossRef]
- Ricart, B.G.; John, B.; Lee, D.; Hunter, C.A.; Hammer, D.A. Dendritic Cells Distinguish Individual Chemokine Signals through CCR7 and CXCR4. J. Immunol. 2010, 186, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, J.; Bierbaum, V.; Merrin, J.; Frank, T.; Hauschild, R.; Bollenbach, T.; Tay, S.; Sixt, M.; Mehling, M. A microfluidic device for measuring cell migration towards substrate-bound and soluble chemokine gradients. Sci. Rep. 2016, 6, 36440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nibbs, R.J.; Graham, G.J. Immune regulation by atypical chemokine receptors. Nat. Rev. Immunol. 2013, 13, 815–829. [Google Scholar] [CrossRef] [PubMed]
- Bachelerie, F.; Ben-Baruch, A.; Burkhardt, A.M.; Combadiere, C.; Farber, J.M.; Graham, G.J.; Horuk, R.; Sparre-Ulrich, A.H.; Locati, M.; Luster, A.D.; et al. International Union of Pharmacology. LXXXIX. Update on the Extended Family of Chemokine Receptors and Introducing a New Nomenclature for Atypical Chemokine Receptors. Pharmacol. Rev. 2014, 66, 1–79. [Google Scholar] [CrossRef] [PubMed]
- Comerford, I.; Milasta, S.; Morrow, V.; Milligan, G.; Nibbs, R. The chemokine receptor CCX-CKR mediates effective scavenging of CCL19 in vitro. Eur. J. Immunol. 2006, 36, 1904–1916. [Google Scholar] [CrossRef] [PubMed]
- Ulvmar, M.H.; Werth, K.; Braun, A.; Kelay, P.; Hub, E.; Eller, K.; Chan, L.; Lucas, B.; Novitzky-Basso, I.; Nakamura, K.; et al. The atypical chemokine receptor CCRL1 shapes functional CCL21 gradients in lymph nodes. Nat. Immunol. 2014, 15, 623–630. [Google Scholar] [CrossRef]
- Moepps, B.; Thelen, M. Monitoring Scavenging Activity of Chemokine Receptors. Methods Enzymol. 2016, 570, 87–118. [Google Scholar]
- Veldkamp, C.T.; Koplinski, C.A.; Jensen, D.R.; Peterson, F.C.; Smits, K.M.; Smith, B.L.; Johnson, S.K.; Lettieri, C.; Buchholz, W.G.; Solheim, J.C.; et al. Production of Recombinant Chemokines and Validation of Refolding. Methods Enzymol. 2016, 570, 539–565. [Google Scholar] [Green Version]
- Otero, C.; Eisele, P.S.; Schaeuble, K.; Groettrup, M.; Legler, D.F. Distinct motifs in the chemokine receptor CCR7 regulate signal transduction, receptor trafficking and chemotaxis. J. Cell Sci. 2008, 121, 2759–2767. [Google Scholar] [CrossRef] [Green Version]
- Uetz-von Allmen, E.; Rippl, A.V.; Farhan, H.; Legler, D.F. A unique signal sequence of the chemokine receptor CCR7 promotes package into COPII vesicles for efficient receptor trafficking. J. Leukocyte Biol. 2018, 104, 375–389. [Google Scholar] [CrossRef]
- Willimann, K.; Legler, D.F.; Loetscher, M.; Roos, R.S.; Delgado, M.B.; Clark-Lewis, I.; Baggiolini, M.; Moser, B. The chemokine SLC is expressed in T cell areas of lymph nodes and mucosal lymphoid tissues and attracts activated T cells via CCR7. Eur. J. Immunol. 1998, 28, 2025–2034. [Google Scholar] [CrossRef] [Green Version]
- Volpe, S.; Cameroni, E.; Moepps, B.; Thelen, S.; Apuzzo, T.; Thelen, M. CCR2 Acts as Scavenger for CCL2 during Monocyte Chemotaxis. PLoS ONE 2012, 7, e37208. [Google Scholar] [CrossRef] [PubMed]
- Schachtner, H.; Weimershaus, M.; Stache, V.; Plewa, N.; Legler, D.F.; Hopken, U.E.; Maritzen, T. Loss of Gadkin Affects Dendritic Cell Migration In Vitro. PLoS ONE 2015, 10, e0143883. [Google Scholar] [CrossRef] [PubMed]
- Hamdan, F.F.; Rochdi, M.D.; Breton, B.; Fessart, D.; Michaud, D.E.; Charest, P.G.; Laporte, S.A.; Bouvier, M. Unraveling G protein-coupled receptor endocytosis pathways using real-time monitoring of agonist-promoted interaction between β-arrestins and AP-2. J. Biol. Chem. 2007, 282, 29089–29100. [Google Scholar] [CrossRef] [PubMed]






© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Purvanov, V.; Matti, C.; Samson, G.P.B.; Kindinger, I.; Legler, D.F. Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions. Int. J. Mol. Sci. 2018, 19, 3876. https://doi.org/10.3390/ijms19123876
Purvanov V, Matti C, Samson GPB, Kindinger I, Legler DF. Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions. International Journal of Molecular Sciences. 2018; 19(12):3876. https://doi.org/10.3390/ijms19123876
Chicago/Turabian StylePurvanov, Vladimir, Christoph Matti, Guerric P. B. Samson, Ilona Kindinger, and Daniel F. Legler. 2018. "Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions" International Journal of Molecular Sciences 19, no. 12: 3876. https://doi.org/10.3390/ijms19123876
APA StylePurvanov, V., Matti, C., Samson, G. P. B., Kindinger, I., & Legler, D. F. (2018). Fluorescently Tagged CCL19 and CCL21 to Monitor CCR7 and ACKR4 Functions. International Journal of Molecular Sciences, 19(12), 3876. https://doi.org/10.3390/ijms19123876