Genome-Wide Identification of microRNAs in Response to Salt/Alkali Stress in Medicago truncatula through High-Throughput Sequencing
Abstract
:1. Introduction
2. Results
2.1. Characterization of Medicago truncatula miRNA Expression in Conditions of Stress
2.2. miRNA Sequencing and Identification
2.3. Clustering Analysis and Identification of Salt/Alkali-Stress-Responsive miRNA
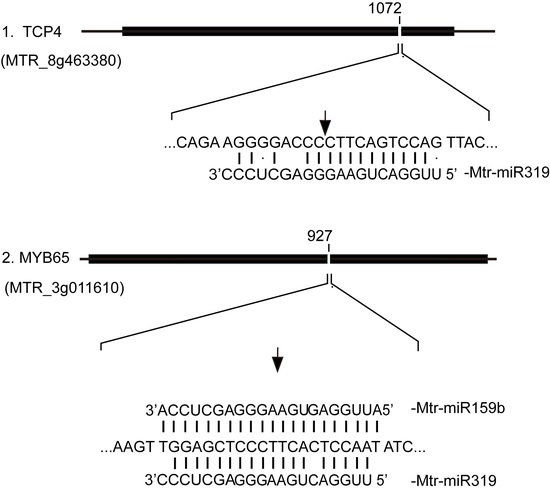
2.4. Prediction and Annotation of Target Genes for Salt/Alkali-Responsive miRNAs
2.5. Validation of High-Throughput Sequencing Data by RT-qPCR and 5’ RACE
3. Discussion
4. Materials and Methods
4.1. Plant Growth and Treatment
4.2. Total RNA Isolation and Sequencing
4.3. Identification of Known and Novel MicroRNAs
4.4. miRNA Target Gene Prediction and Function Analysis
4.5. miRNA Validation by RT-qPCR
4.6. Detection of miRNA Cleavage Targets
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Rockström, J.; Steffen, W.; Noone, K.; Persson, A.; Rd, C.F.; Lambin, E.F.; Lenton, T.M.; Scheffer, M.; Folke, C.; Schellnhuber, H.J. A safe operating space for humanity. Nature 2009, 461, 472–475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef]
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, B. MicroRNA: A new target for improving plant tolerance to abiotic stress. J. Exp. Bot. 2015, 66, 1749–1761. [Google Scholar] [CrossRef]
- Barciszewskapacak, M.; Milanowska, K.; Knop, K.; Bielewicz, D.; Nuc, P.; Plewka, P.; Pacak, A.M.; Vazquez, F.; Karlowski, W.; Jarmolowski, A. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front. Plant Sci. 2015, 6, 410. [Google Scholar] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Pratt, A.J.; Macrae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, physiology, and molecular breeding approaches for improving salt tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef] [PubMed]
- Mantri, N.; Basker, N.; Ford, R. The role of miRNAs in legumes with a focus on abiotic stress response. Plant Genome 2015, 6, 841–856. [Google Scholar]
- Ding, Y.; Tao, Y.; Zhu, C. Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 2013, 64, 3077–3086. [Google Scholar] [CrossRef] [Green Version]
- Ferdous, J.; Hussain, S.S.; Shi, B.J. Role of microRNAs in plant drought tolerance. Plant Biotechnol. J. 2015, 13, 293–305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Syuhada, O.N.; Kalaivani, N. Investigating the roles of MicroRNAs in biotic stress response induced by Rhizoctonia solani in rice. In Proceedings of the Ukm Fst Postgraduate Colloquium: Universiti Kebangsaan Malaysia, Faculty of Science & Technology Postgraduate Colloquium, Selangor, Malaysia, 9–11 April 2014; pp. 761–764. [Google Scholar]
- Li, W.; Wang, T.; Zhang, Y.; Li, Y. Overexpression of soybean miR172c confers tolerance to water deficit and salt stress, but increases ABA sensitivity in transgenic Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 175–194. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, D.; Li, Z.; Hu, Q.; Yang, C.; Zhu, L.; Luo, H. Constitutive expression of a miR319 gene alters plant development and enhances salt and drought tolerance in transgenic creeping bentgrass. Plant Physiol. 2013, 161, 1375–1391. [Google Scholar] [CrossRef] [PubMed]
- Yuan, S.; Li, Z.; Li, D.; Yuan, N.; Hu, Q.; Luo, H. Constitutive expression of rice microRNA528 alters plant development and enhances tolerance to salinity stress and nitrogen starvation in creeping bentgrass. Plant Physiol. 2015, 169, 576–593. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Yang, L.; Zeng, H.Q.; Zhou, Z.S.; Yang, Z.M.; Li, H.; Sun, D.; Xie, Fs.; Zhang, B. A cotton miRNA is involved in regulation of plant response to salt stress. Sci. Rep. 2016, 6, 19736. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Zheng, H.; Jiang, Q.; Hui, Z. GmNFYA3, a target gene of miR169, is a positive regulator of plant tolerance to drought stress. Plant Mol. Biol. 2013, 82, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Luan, M.; Xu, M.; Lu, Y.; Zhang, L.; Fan, Y.; Wang, L. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 555, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Ge, L.; Liang, R.; Wei, L.; Ruan, K.; Lin, H.; Jin, Y. Members of miR-169 family are induced by high salinity and transiently inhibit the NF-YA transcription factor. BMC Mol. Biol. 2009, 10, 29. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant 2011, 143, 1–9. [Google Scholar] [CrossRef]
- Graham, P.H.; Vance, C.P. Legumes: Importance and constraints to greater use. Plant Physiol. 2003, 131, 872–877. [Google Scholar] [CrossRef]
- Islam, M.R.; Salam, M.A.; Bhuiyan, M.A.R.; Rahman, M.A.; Yasmeen, R.; Rahman, M.S.; Uddin, M.K.; Gregorio, G.B.; Ismail, A.M. A salt tolerant rice variety for boro season isolated through participatory variety selection. Int. J. Robot. Res. 2008, 5, 1–6. [Google Scholar]
- Parida, A.K.; Das, A.B. Salt tolerance and salinity effects on plants: A review. Ecotoxicol. Environ. Saf. 2005, 60, 324–349. [Google Scholar] [CrossRef]
- Aung, B.; Gruber, M.Y.; Amyot, L.; Omari, K.; Bertrand, A.; Hannoufa, A. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 2015, 13, 779–790. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Gruber, M.Y.; Wall, K.; Hannoufa, A. An Insight into microRNA156 Role in salinity stress responses of alfalfa. Front. Plant Sci. 2017, 8, 356. [Google Scholar] [CrossRef] [PubMed]
- Li, R.L.; Shi, F.C.; Fukuda, K.; Yang, Y.L. Effects of salt and alkali stresses on germination, growth, photosynthesis and ion accumulation in alfalfa (Medicago sativa L.). Soil Sci. Plant Nutr. 2010, 56, 725–733. [Google Scholar] [CrossRef]
- Sun, X.; Xu, L.; Wang, Y.; Yu, R.; Zhu, X.; Luo, X.; Gong, Y.; Wang, R.; Limera, C.; Zhang, K.; et al. Identification of novel and salt-responsive miRNAs to explore miRNA-mediated regulatory network of salt stress response in radish (Raphanus sativus L.). BMC Genom. 2015, 16, 197. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.N.; Li, X.; Liu, J.H. Identification of conserved and novel cold-responsive microRNAs in trifoliate orange (Poncirus trifoliata (L.) Raf.) using high-throughput sequencing. Plant Mol. Biol. Rep. 2014, 32, 328–341. [Google Scholar] [CrossRef]
- Ren, Y.; Chen, L.; Zhang, Y.; Kang, X.; Zhang, Z.; Wang, Y. Identification and characterization of salt-responsive microRNAs in Populus tomentosa by high-throughput sequencing. Biochimie 2013, 95, 743–750. [Google Scholar] [CrossRef]
- Pantaleo, V.; Szittya, G.; Moxon, S.; sMiozzi, L.; Moulton, V.; Dalmay, T.; Burgyan, J. Identification of grapevine microRNAs and their targets using high-throughput sequencing and degradome analysis. Plant J. 2010, 62, 960–976. [Google Scholar]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef]
- Yang, X.; Li, L. miRDeep-P: A computational tool for analyzing the microRNA transcriptome in plants. Bioinformatics 2011, 27, 2614–2615. [Google Scholar] [CrossRef] [PubMed]
- Allen, E.; Xie, Z.; Gustafson, A.M.; Carrington, J.C. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 2005, 121, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Llave, C.; Xie, Z.; Kasschau, K.D.; Carrington, J.C. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 2002, 297, 2053–2056. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Liu, Z.; Dai, X.; Xiang, F. Primary root growth in Arabidopsis thaliana is inhibited by the miR159 mediated repression of MYB33, MYB65 and MYB101. Plant Sci. 2017, 262, 182–189. [Google Scholar]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2017, 176, 1694–1708. [Google Scholar] [CrossRef] [PubMed]
- Larcher, W. Physiological Plant Ecology; Springer-Verlag: Berlin, Germany, 2003; Volume 32, pp. 1217–1220. [Google Scholar]
- Bahmani, K.; Noori, S.A.S.; Darbandi, A.I.; Akbari, A. Molecular mechanisms of plant salinity tolerance: A review. Aust. J. Crop Sci. 2015, 9, 321–336. [Google Scholar]
- Long, R.C.; Li, M.N.; Kang, J.M.; Zhang, T.J.; Sun, Y.; Yang, Q.C. Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula. Physiol. Plant 2015, 154, 13–27. [Google Scholar] [CrossRef]
- Li, Y.; Wan, L.; Bi, S.; Wan, X.; Li, Z.; Cao, J.; Tong, Z.; Xu, H.; He, F.; Li, X. Identification of drought-responsive microRNAs from roots and leaves of alfalfa by high-throughput sequencing. Genes 2017, 8, 119. [Google Scholar] [CrossRef]
- Kawashima, C.G.; Matthewman, C.A.; Huang, S.; Lee, B.R.; Yoshimoto, N.; Koprivova, A.; Rubio-Somoza, I.; Todesco, M.; Rathjen, T.; Saito, K. Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 2011, 66, 863–876. [Google Scholar] [CrossRef] [Green Version]
- Trindade, I.; Capitao, C.; Dalmay, T.; Fevereiro, M.P.; Santos, D.M. miR398 and miR408 are up-regulated in response to water deficit in Medicago truncatula. Planta 2010, 231, 705–716. [Google Scholar] [CrossRef]
- Jia, X.; Wang, W.X.; Ren, L.; Chen, Q.J.; Mendu, V.; Willcut, B.; Dinkins, R.; Tang, X.; Tang, G. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol. Biol. 2009, 71, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Burd, S.; Lers, A. miR408 is involved in abiotic stress responses in Arabidopsis. Plant J. 2015, 84, 169–187. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Li, Y.; Cai, H.; Ji, W.; Guo, D.; Zhu, Y. Over-expression of osa—miR396c decreases salt and alkali stress tolerance. Planta 2010, 231, 991–1001. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.; Wang, R.; Ou, X.; Fang, Z.; Tian, C.; Duan, J.; Wang, Y.; Zhang, M. OsTIR1 and OsAFB2 downregulation via OsmiR393 overexpression leads to more tillers, early flowering and less tolerance to salt and drought in rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J.; Meyers, B.C. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell 2018, 30, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Bischof, S.; Wang, H.; Feng, S.; Lee, T.F.; Teng, C.; Chen, X.; Park, S.Y.; Liu, L.; Gallego-Bartolome, J. A one precursor one siRNA model for pol IV-dependent siRNA biogenesis. Cell 2015, 163, 445–455. [Google Scholar] [CrossRef]
- Meyers, B.C.; Axtell, M.J.; Bonnie, B.; Bartel, D.P.; David, B.; Bowman, J.L.; Xiaofeng, C.; Carrington, J.C.; Xuemei, C.; Green, P.J. Criteria for annotation of plant microRNAs. Plant Cell 2008, 20, 3186–3190. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Wu, G.; Park, M.Y.; Conway, S.R.; Wang, J.W.; Weigel, D.; Poethig, R.S. The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 2009, 138, 750–759. [Google Scholar] [CrossRef]
- Yu, S.; Cao, L.; Zhou, C.M.; Zhang, T.Q.; Lian, H.; Sun, Y.; Huang, J.; Wang, G.; Wang, J.W. Sugar is an endogenous cue for juvenile-to-adult phase transition in plants. Elife 2013, 2, e00269. [Google Scholar] [CrossRef]
- Buendía-Monreal, M.; Gillmor, C.S. Convergent repression of miR156 by sugar and the CDK8 module of Arabidopsis Mediator. Dev. Biol. 2017, 423, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Wen, D.; Huang, L.; Song, A.; Wang, H.; Fang, W.; Chen, F.; Teng, N. Identification of microRNAs and their targets associated with embryo abortion during chrysanthemum cross breeding via high-throughput sequencing. PLoS ONE 2015, 10, e0124371. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Chen, Y.Q. The potential roles of microRNAs in molecular breeding. Methods Mol. Biol. 2012, 877, 303–311. [Google Scholar] [PubMed]
- Tang, J.; Chu, C. MicroRNAs in crop improvement: Fine-tuners for complex traits. Nat. Plants 2017, 3, 17077. [Google Scholar] [CrossRef] [PubMed]
- Si-Ammour, A.; Windels, D.; Arn-Bouldoires, E.; Kutter, C.; Ailhas, J.; Meins, F.; Vazquez, F. miR393 and secondary siRNAs regulate expression of the TIR1/AFB2 auxin receptor clade and auxin-related development of Arabidopsis leaves. Plant Physiol. 2011, 157, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Bai, X.; Yang, L.; Lv, D.; Pan, X.; Li, Y.; Cai, H.; Ji, W.; Chen, Q.; Zhu, Y. osa-miR393: A salinity- and alkaline stress-related microRNA gene. Mol. Biol. Rep. 2011, 38, 237–242. [Google Scholar] [CrossRef]
- Chen, Z.; Hu, L.; Han, N.; Hu, J.; Yang, Y.; Xiang, T.; Zhang, X.; Wang, L. Overexpression of a miR393-resistant form of transport inhibitor response protein 1 (mTIR1) enhances salt tolerance by increased osmoregulation and Na+ exclusion in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Bonnet, E.; Wuyts, J.; Rouzé, P.; Peer, Y.V.D. Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 2004, 20, 2911–2917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, Y.; Jianqi, L.I.; Songfeng, W.U.; Zhu, Y.; Chen, Y.; Fuchu, H.E. Integrated NR annotation system and its localization. Comput. Eng. 2006, 32, 71–72. [Google Scholar]










| Library | Repetition | Total Reads | Clean Reads | Reads Mapped to the Genome | Known-miRNAs | Novel-miRNAs |
|---|---|---|---|---|---|---|
| DW | DW1 | 43,687,837 | 39,659,762 | 13,087,074 | 575 | 202 |
| DW2 | 33,719,419 | 31,757,315 | 11,898,180 | 587 | 205 | |
| DW3 | 31,320,157 | 29,377,694 | 11,744,041 | 589 | 206 | |
| SS | SS1 | 32,316,320 | 29,294,137 | 10,926,133 | 568 | 201 |
| SS2 | 28,977,350 | 24,356,163 | 9,579,601 | 568 | 205 | |
| SS3 | 56,322,861 | 51,774,436 | 19,162,589 | 594 | 204 | |
| AS | AS1 | 39,385,384 | 31,654,898 | 13,191,666 | 586 | 204 |
| AS2 | 44,183,950 | 39,649,487 | 14,993,557 | 586 | 201 | |
| AS3 | 36,268,593 | 32,538,226 | 10,722,784 | 581 | 205 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, C.; Long, R.; Zhang, T.; Kang, J.; Wang, Z.; Wang, P.; Sun, H.; Yu, J.; Yang, Q. Genome-Wide Identification of microRNAs in Response to Salt/Alkali Stress in Medicago truncatula through High-Throughput Sequencing. Int. J. Mol. Sci. 2018, 19, 4076. https://doi.org/10.3390/ijms19124076
Cao C, Long R, Zhang T, Kang J, Wang Z, Wang P, Sun H, Yu J, Yang Q. Genome-Wide Identification of microRNAs in Response to Salt/Alkali Stress in Medicago truncatula through High-Throughput Sequencing. International Journal of Molecular Sciences. 2018; 19(12):4076. https://doi.org/10.3390/ijms19124076
Chicago/Turabian StyleCao, Chunyu, Ruicai Long, Tiejun Zhang, Junmei Kang, Zhen Wang, Pingqing Wang, Hao Sun, Jie Yu, and Qingchuan Yang. 2018. "Genome-Wide Identification of microRNAs in Response to Salt/Alkali Stress in Medicago truncatula through High-Throughput Sequencing" International Journal of Molecular Sciences 19, no. 12: 4076. https://doi.org/10.3390/ijms19124076
APA StyleCao, C., Long, R., Zhang, T., Kang, J., Wang, Z., Wang, P., Sun, H., Yu, J., & Yang, Q. (2018). Genome-Wide Identification of microRNAs in Response to Salt/Alkali Stress in Medicago truncatula through High-Throughput Sequencing. International Journal of Molecular Sciences, 19(12), 4076. https://doi.org/10.3390/ijms19124076