Lipid Supplement in the Cultural Condition Facilitates the Porcine iPSC Derivation through cAMP/PKA/CREB Signal Pathway
Abstract
:1. Introduction
2. Results
2.1. Fatty Acid Enriched Culture Condition Improves Reprogramming Efficiency
2.2. The iPSCs Have Pluripotency, Differentiation Potential and Embryo Aggregation Abilities
2.3. AlbuMAX Promotes iPSCs Proliferation
2.4. AlbuMAX Contributes to Lipid Droplets Accumulation
2.5. AlbuMAX Promotes MET During the Reprogramming of Fibroblasts
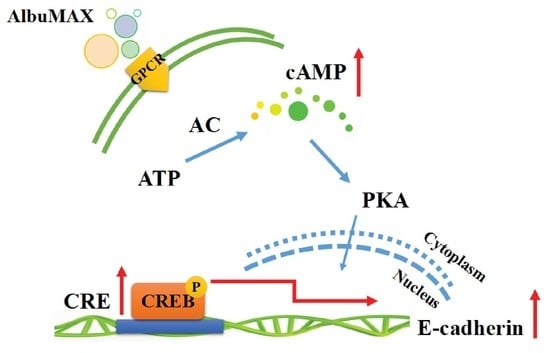
2.6. AlbuMAX Enhances Reprogramming via Activating cAMP/PKA/CREB Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Animal Experiments
4.2. Generation of Porcine iPSCs and Cell Culture
4.3. Quantitative RT-PCR
| Forward (5′ to 3′) | Reverse (5′ to 3′) | References | |
| pMXs-Oct4 | GACGGCATCGCAGCTTGGATACAC | GAGAAGGCGAAGTCGGAAG | [29] |
| pMXs-Sox2 | GACGGCATCGCAGCTTGGATACAC | GGCTGTTCTTCTGGTTGC | [29] |
| pMXs-Klf4 | GACGGCATCGCAGCTTGGATACAC | GTCTTTGCTTCATGTGGG | [29] |
| pMXs-Myc | GACGGCATCGCAGCTTGGATACAC | GAAATAAGGCTGCACCGAGT | [29] |
| Nanog | CATCTGCTGAGACCCTCGAC | GGGCTTGTGGAAGAATCAGG | [29] |
| EF-1α | AATGCGGTGGGATCGACAAA | CACGCTCACGTTCAGCCTTT | [29] |
| Endo-Oct4 | CAAACTGAGGTGCCTGCCCTTC | CAAACTGAGGTGCCTGCCCTTC | [29] |
| Endo-Sox2 | CATCAACGGTACACTGCCTCTC | ACTCTCCTCCCATTTCCCTCTTT | [29] |
| Endo-Klf4 | CATGAGTTGGGGGAGGGAAG | ACTCACCAAGCACCATCGTT | [29] |
| Endo-Myc | ATCCAAGACCACCACCACTG | ATCCAAGACCACCACCACTG | [29] |
| Cdh1 | TGGGCCGAGTGAGTTTTGAA | TGACTGTAACCACACCGTCG | [29] |
| Sall4 | ATCGACGTTTATCCGAGCCC | ATCGACGTTTATCCGAGCCC | [29] |
| EpCAM | TGCTCTTTGAATGCGCTTGG | AGAGCCCATCGTTGTTCTGG | |
| Esrrb | CCGGACAAACTCTACGCCAT | GCTTGGCCCAACCAATGATG | |
| Cdc2 | TTTTCAAAGCTGGCTCTGGGAG | GGATGTGGTAGATCCCAGCTTA | |
| Ccnc | AGAAAGATGCCAGACGGTGG | AGGAGGTTTTGGTTTCGGCA | |
| Ccnb1 | AGATCGCAGCAGGAGCTTTT | CCTCGATTCACCACGACGAT | |
| Ccna2 | CTAACATTGCAGCAGACGGC | ATCCTTAAGAGGCGCAACCC | |
| Acox2 | AGGACTCAGGACGAGACACA | TTGAAGGACGGCATGCATCT | |
| Prkaa2 | ATTCTGTCACTGCGGAGAGC | AATCCATGGTGTGACTGCCC | |
| Acadvl | GAAGTCAAATGCCTGCCAGC | ATGTTGGCGCTCACCATGTA | |
| Acadsb | ACACCAAGTGGCTCATACGG | TACCAATCTTCGCGTCTCGG | |
| Fabp5 | AGGCACCAGTCCGCTTATTC | TTTCGTAGGGCCATTCCCAC |
4.4. Alkaline Phosphatase Staining and Immunofluorescence Staining
4.5. Flow Cytometry Analysis
4.6. Teratoma Formation Assay
4.7. Parthenogenetic Embryo Injection
4.8. Western Blot
4.9. Detection of cAMP Levels
4.10. The Luciferase Reporter Construction and Luciferase Activity Analysis
| Top chain | tcgagACTCTGTCTCAGTTATTTTTCCCTCGTCAAGAGCCATCTGAAGGAGAGGCa |
| Bottom chain | agcttGCCTCTCCTTCAGATGGCTCTTGACGAGGGAAAAATAACTGAGACAGAGTc |
| Forward | AGTTATTTTTCCCTCTGCAAGAGCCATCT |
| Reverse | AGATGGCTCTTGCAGAGGGAAAAATAACT |
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiue, Y.L.; Yang, J.R.; Liao, Y.J.; Kuo, T.Y.; Liao, C.H.; Kang, C.H.; Tai, C.; Anderson, G.B.; Anderson, L. Derivation of porcine pluripotent stem cells for biomedical research. Theriogenology 2016, 86, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Han, J.; Wu, J.; Li, Q.; Liu, S.; Zhang, W.; Pei, Y.; Ruan, X.; Liu, Z.; Wang, X.; et al. Specific gene-regulation networks during the pre-implantation development of the pig embryo as revealed by deep sequencing. BMC Genom. 2014, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Montserrat, N.; Bahima, E.G.; Batlle, L.; Hafner, S.; Rodrigues, A.M.; Gonzalez, F.; Belmonte, J.C.I. Generation of pig iPS cells: A model for cell therapy. J. Cardiovasc. Transl. Res. 2011, 4, 121–130. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, Q.; Deng, W.; Wang, H.; Liu, K.; Fu, H.; Zhao, Q.; Wang, X.; Liu, L. Epigenetic modifiers facilitate induction and pluripotency of porcine iPSCs. Stem Cell Rep. 2017, 8, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Petkov, S.; Glage, S.; Nowak-Imialek, M.; Niemann, H. Long-Term Culture of Porcine Induced Pluripotent Stem-Like Cells Under Feeder-Free Conditions in the Presence of Histone Deacetylase Inhibitors. Stem Cells Dev. 2016, 25, 386–394. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Park, J.K.; Son, D.; Hwang, J.Y.; Lee, D.K.; Ka, H.; Park, J.; Lee, C. Reactivation of endogenous genes and epigenetic remodeling are barriers for generating transgene-free induced pluripotent stem cells in pig. PLoS ONE 2016, 11, e0158046. [Google Scholar] [CrossRef] [PubMed]
- Sturmey, R.G.; Reis, A.; Leese, H.J.; McEvoy, T.G. Role of fatty acids in energy provision during oocyte maturation and early embryo development. Reprod. Domest Anim. 2009, 44 (Suppl. 3), 50–58. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.H.; Kim, M.O.; Kim, Y.H.; Kim, J.S.; Han, H.J. Linoleic acid induces mouse embryonic stem cell proliferation via Ca2+/PKC, PI3K/Akt, and MAPKs. Cell Physiol. Biochem. 2009, 23, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Hejna, M.; Liu, Y.; Percharde, M.; Wossidlo, M.; Blouin, L.; Durruthy-Durruthy, J.; Wong, P.; Qi, Z.; Yu, W.; et al. YAP Induces Human Naive Pluripotency. Cell Rep. 2016, 14, 2301–2312. [Google Scholar] [CrossRef] [PubMed]
- Pebay, A.; Wong, R.C.; Pitson, S.M.; Wolvetang, E.J.; Peh, G.S.; Filipczyk, A.; Koha, K.L.L.; Tellisa, I.; Nguyena, L.T.V.; Peraa, M.F. Essential roles of sphingosine-1-phosphate and platelet-derived growth factor in the maintenance of human embryonic stem cells. Stem Cells 2005, 23, 1541–1548. [Google Scholar] [CrossRef] [PubMed]
- Pitson, S.M.; Pebay, A. Regulation of stem cell pluripotency and neural differentiation by lysophospholipids. Neurosignals. 2009, 17, 242–254. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, C.; Sakaguchi, Y.; Hoshi, H.; Yoshioka, K. Lipid-rich bovine serum albumin improves the viability and hatching ability of porcine blastocysts produced in vitro. J. Reprod. Dev. 2016, 62, 79–86. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gonzalo, F.R.; Izpisua Belmonte, J.C. Albumin-associated lipids regulate human embryonic stem cell self-renewal. PLoS ONE 2008, 3, e1384. [Google Scholar] [CrossRef] [PubMed]
- Rajanahalli, P.; Meyer, K.; Zhu, L.; Wagner, B.D.; Robinson, M.L.; King, D.A.; Hong, Y. Conversion of mouse fibroblasts to sphere cells induced by AlbuMAXI-containing medium. Front. Biosci. 2012, 4, 1813–1822. [Google Scholar] [CrossRef]
- Gafni, O.; Weinberger, L.; Mansour, A.A.; Manor, Y.S.; Chomsky, E.; Ben-Yosef, D.; Kalma, Y.; Viukov, S.; Maza, I.; Zviran, A.; et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013, 504, 282–286. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Chen, K.S.; Zeng, X.M.; Yang, J.G.; Wu, Y.; Shi, X.; Qin, B.; Zeng, L.; Esteban, M.A.; Pan, D.; et al. The histone demethylases jhdm1a/1b enhance somatic cell reprogramming in a vitamin-c-dependent manner. Cell Stem Cell 2011, 9, 575–587. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, S.; Panopoulos, A.D.; Herrerias, A.; Bissig, K.D.; Lutz, M.; Berggren, W.T.; Verma, I.M.; Belmonte, J.C.I. A high proliferation rate is required for cell reprogramming and maintenance of human embryonic stem cell identity. Current Biol. 2011, 21, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Plath, K.; Lowry, W.E. Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 2011, 12, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Guo, Y.; Cui, Y.; Liu, Y.; Yu, T.; Wang, H. Generation of intermediate porcine iPS cells under culture condition favorable for mesenchymal-to-epithelial transition. Stem Cell Rev. 2015, 11, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Binder, B.Y.; Williams, P.A.; Silva, E.A.; Leach, J.K. Lysophosphatidic acid and sphingosine-1-phosphate: A concise review of biological function and applications for tissue engineering. Tissue Eng. Part B Rev. 2015, 21, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Huangfu, D.W.; Osafune, K.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Chen, S.; Muhlestein, W.; Melton, D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008, 26, 1269–1275. [Google Scholar]
- Kim, J.S.; Chae, J.I.; Song, B.S.; Lee, K.S.; Choo, Y.K.; Chang, K.T.; Park, H.; Koo, D.B. Iloprost, a prostacyclin analogue, stimulates meiotic maturation and early embryonic development in pigs. Reprod. Fertil. Dev. 2010, 22, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Redmer, T.; Diecke, S.; Grigoryan, T.; Quiroga-Negreira, A.; Birchmeier, W.; Besser, D. E-cadherin is crucial for embryonic stem cell pluripotency and can replace OCT4 during somatic cell reprogramming. EMBO Rep. 2011, 12, 720–726. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Yuan, D.; Wei, B.; Jiang, J.; Kang, J.; Ling, K.; Gu, Y.; Li, J.; Xiao, L.; Pei, G. E-cadherin-mediated cell-cell contact is critical for induced pluripotent stem cell generation. Stem Cells. 2010, 28, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Han, J.; Li, P.; Cao, S.; An, Y.; Lim, B.; Li, N. A novel strategy to derive iPS cells from porcine fibroblasts. Sci. China Life Sci. 2011, 54, 553–559. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Pei, Y.; Zhong, L.; Wen, B.; Cao, S.; Han, J. Pluripotent and metabolic features of two types of porcine iPSCs derived from defined mouse and human es cell culture conditions. PLoS ONE 2015, 10, e0124562. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Zhou, N.; Zhang, Y.; Zhang, Y.; Wu, R.; Li, Y.; Zhang, Y.; Li, N. Dynamic reprogramming of 5-hydroxymethylcytosine during early porcine embryogenesis. Theriogenology 2014, 81, 496–508. [Google Scholar] [CrossRef] [PubMed]







© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, W.; Wang, H.; Zhang, S.; Zhong, L.; Wang, Y.; Pei, Y.; Han, J.; Cao, S. Lipid Supplement in the Cultural Condition Facilitates the Porcine iPSC Derivation through cAMP/PKA/CREB Signal Pathway. Int. J. Mol. Sci. 2018, 19, 509. https://doi.org/10.3390/ijms19020509
Zhang W, Wang H, Zhang S, Zhong L, Wang Y, Pei Y, Han J, Cao S. Lipid Supplement in the Cultural Condition Facilitates the Porcine iPSC Derivation through cAMP/PKA/CREB Signal Pathway. International Journal of Molecular Sciences. 2018; 19(2):509. https://doi.org/10.3390/ijms19020509
Chicago/Turabian StyleZhang, Wei, Hanning Wang, Shaopeng Zhang, Liang Zhong, Yanliang Wang, Yangli Pei, Jianyong Han, and Suying Cao. 2018. "Lipid Supplement in the Cultural Condition Facilitates the Porcine iPSC Derivation through cAMP/PKA/CREB Signal Pathway" International Journal of Molecular Sciences 19, no. 2: 509. https://doi.org/10.3390/ijms19020509
APA StyleZhang, W., Wang, H., Zhang, S., Zhong, L., Wang, Y., Pei, Y., Han, J., & Cao, S. (2018). Lipid Supplement in the Cultural Condition Facilitates the Porcine iPSC Derivation through cAMP/PKA/CREB Signal Pathway. International Journal of Molecular Sciences, 19(2), 509. https://doi.org/10.3390/ijms19020509