AP-1/KIF13A Blocking Peptides Impair Melanosome Maturation and Melanin Synthesis
Abstract
:1. Introduction
2. Results
2.1. β1-Adaptin-Derived Peptides Prevent the AP-1/KIF13A Interaction
2.2. β1-Adaptin-Derived Peptides Displace the AP-1/KIF13A Co-Distribution
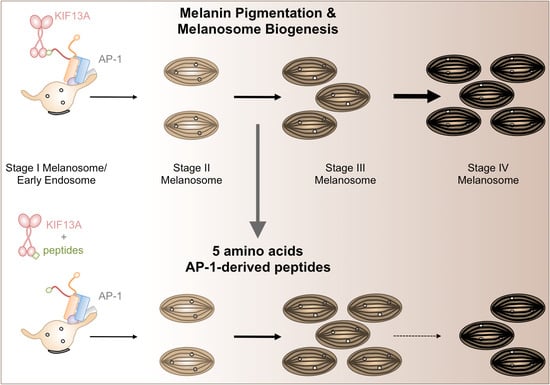
2.3. β1-Adaptin-Derived Peptides Decrease Pigmentation And Melanosome Biogenesis
2.4. β1-Adaptin-Derived Peptides Might Confer Skin Hypopigmentation
3. Discussion
4. Materials and Methods
4.1. Peptide Design, Synthesis, and Preparation
4.2. Cell Culture
4.3. Protein Extraction, Immunoprecipitation, and Western Blot Analysis
4.4. Immunofluorescence
4.5. Co-Localization Analysis
4.6. Conventional Electron Microscopy
4.7. Melanin Assay
4.8. Statistical Analysis
Acknowledgments
Author contributions
Conflicts of Interest
Abbreviations
| TYRP1 | tyrosinase-related protein 1 |
| AP-1 | adaptor protein-1 |
| aa | amino acid |
References
- Seiji, M.; Fitzpatrick, T.M.; Simpson, R.T.; Birbeck, M.S.C. Chemical composition and terminology of specialized organelles (melanosomes and melanin granules) in mammalian melanocytes. Nature 1963, 197, 1082–1084. [Google Scholar] [CrossRef] [PubMed]
- Hurbain, I.; Geerts, W.J.; Boudier, T.; Marco, S.; Verkleij, A.J.; Marks, M.S.; Raposo, G. Electron tomography of early melanosomes: Implications for melanogenesis and the generation of fibrillar amyloid sheets. Proc. Natl. Acad. Sci. USA 2008, 105, 19726–19731. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Tenza, D.; Murphy, D.M.; Berson, J.F.; Marks, M.S. Distinct protein sorting and localization to premelanosomes, melanosomes, and lysosomes in pigmented melanocytic cells. J. Cell Biol. 2001, 152, 809–824. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Hammer, J.A. Melanosome transfer: It is best to give and receive. Curr. Opin. Cell Biol. 2014, 29, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Sitaram, A.; Marks, M.S. Mechanisms of protein delivery to melanosomes in pigment cells. Physiology 2012, 27, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Delevoye, C.; Hurbain, I.; Tenza, D.; Sibarita, J.B.; Uzan-Gafsou, S.; Ohno, H.; Geerts, W.J.; Verkleij, A.J.; Salamero, J.; Marks, M.S.; et al. AP-1 and KIF13A coordinate endosomal sorting and positioning during melanosome biogenesis. J. Cell Biol. 2009, 187, 247–264. [Google Scholar] [CrossRef] [PubMed]
- Theos, A.C.; Tenza, D.; Martina, J.A.; Hurbain, I.; Peden, A.A.; Sviderskaya, E.V.; Stewart, A.; Robinson, M.S.; Bennett, D.C.; Cutler, D.F.; et al. Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol. Biol. Cell 2005, 16, 5356–5372. [Google Scholar] [CrossRef] [PubMed]
- Delevoye, C.; Heiligenstein, X.; Ripoll, L.; Gilles-Marsens, F.; Dennis, M.K.; Linares, R.A.; Derman, L.; Gokhale, A.; Morel, E.; Faundez, V.; et al. BLOC-1 brings together the actin and microtubule cytoskeletons to generate recycling endosomes. Curr. Biol. 2016, 26, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Delevoye, C.; Miserey-Lenkei, S.; Montagnac, G.; Gilles-Marsens, F.; Paul-Gilloteaux, P.; Giordano, F.; Waharte, F.; Marks, M.S.; Goud, B.; Raposo, G. Recycling endosome tubule morphogenesis from sorting endosomes requires the kinesin motor KIF13A. Cell Rep. 2014, 6, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, T.; Setou, M.; Seog, D.; Ogasawara, K.; Dohmae, N.; Takio, K.; Hirokawa, N. A novel motor, KIF13A, transports mannose-6-phosphate receptor to plasma membrane through direct interaction with AP-1 complex. Cell 2000, 103, 569–581. [Google Scholar] [CrossRef]
- Ebanks, J.P.; Wickett, R.R.; Boissy, R.E. Mechanisms regulating skin pigmentation: The rise and fall of complexion coloration. Int. J. Mol. Sci. 2009, 10, 4066–4087. [Google Scholar] [CrossRef] [PubMed]
- Solano, F.; Briganti, S.; Picardo, M.; Ghanem, G. Hypopigmenting agents: An updated review on biological, chemical and clinical aspects. Pigment Cell Res. 2006, 19, 550–571. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, K.; Tsukamoto, K.; Okazaki, M.; Virador, V.M.; Lei, T.C.; Suzuki, Y.; Uchida, G.; Kitano, Y.; Harii, K. Effects of all-trans retinoic acid on melanogenesis in pigmented skin equivalents and monolayer culture of melanocytes. J. Dermatol. Sci. 2001, 27, S68–S75. [Google Scholar] [CrossRef]
- Huang, Z.M.; Chinen, M.; Chang, P.J.; Xie, T.; Zhong, L.; Demetriou, S.; Patel, M.P.; Scherzer, R.; Sviderskaya, E.V.; Bennett, D.C.; et al. Targeting protein-trafficking pathways alters melanoma treatment sensitivity. Proc. Natl. Acad. Sci. USA 2012, 109, 553–558. [Google Scholar] [CrossRef] [PubMed]
- Xie, T.; Nguyen, T.; Hupe, M.; Wei, M.L. Multidrug resistance decreases with mutations of melanosomal regulatory genes. Cancer Res. 2009, 69, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Nakatsu, F.; Hase, K.; Ohno, H. The role of the clathrin adaptor AP-1: Polarized sorting and beyond. Membranes 2014, 4, 747–763. [Google Scholar] [CrossRef] [PubMed]
- Campagne, C.; Ripoll, L.; Gilles-Marsens, F.; Raposo, G.; Delevoye, C. Melanin estimation of peptide-treated MNT-1 cells (1 µM and 5 µM peptide concentration). Unpublished work. 2018. [Google Scholar]
- Zhou, R.; Niwa, S.; Guillaud, L.; Tong, Y.; Hirokawa, N. A molecular motor, KIF13A, controls anxiety by transporting the serotonin type 1A receptor. Cell Rep. 2013, 3, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Nascimento, A.; Kellen, B.; Ferreira, F.; Alenquer, M.; Vale-Costa, S.; Raposo, G.; Delevoye, C.; Amorim, M.J. KIF13A mediates trafficking of influenza A virus ribonucleoproteins. J. Cell Sci. 2017, 130, 4038–4050. [Google Scholar] [CrossRef] [PubMed]
- Bechara, C.; Sagan, S. Cell-penetrating peptides: 20 years later, where do we stand? FEBS Lett. 2013, 587, 1693–1702. [Google Scholar] [CrossRef] [PubMed]
- Lo Cicero, A.; Delevoye, C.; Gilles-Marsens, F.; Loew, D.; Dingli, F.; Guere, C.; Andre, N.; Vie, K.; van Niel, G.; Raposo, G. Exosomes released by keratinocytes modulate melanocyte pigmentation. Nat. Commun. 2015, 6, 7506. [Google Scholar] [CrossRef] [PubMed]


| Peptide (Solvent) | 11aa Peptide Sequences | Peptide (Solvent) | 5aa Peptide Sequences | Peptide (Solvent) | 3aa Peptide Sequences |
|---|---|---|---|---|---|
| AI-11 (H2O) | APLSPNQTVEI | QI-5 (H2O) | QTVEI | GV-3 (H2O) | GSV |
| QT-11 (H2O) | QTVEISLPLST | EP-5 (H2O) | EISLP | VK-3 (H2O) | VMK |
| LK-11 (DMSO) | LPLSTVGSVMK | LT-5 (H2O) | LPLST | EL-3 (H2O) | EPL |
| GN-11 (H2O) | GSVMKMEPLNN | SS-5 (H2O) | STVGS | LN-3 (H2O) | LNN |
| EK-11 (H2O) | EPLNNLQVAVK | GK-5 (H2O) | GSVMK | QA-3 (H2O) | QVA |
| QF-11 (DMSO) | QVAVKNNIDVF | MP-5 (H2O) | MKMEP | AK-3 (H2O) | AVK |
| NY-11 (DMSO) | NIDVFYFSTLY | EN-5 (H2O) | EPLNN | ||
| FF-11 (DMSO) | FSTLYPLHILF | NV-5 (H2O) | NNLQV | ||
| LM-11 (DMSO) | LHILFVEDGKM | QK-5 (H2O) | QVAVK | ||
| EL-11 (H2O) | EDGKMDRQMFL | ||||
| RI-11 (DMSO) | RQMFLATWKDI | ||||
| WQ-11 (H2O) | WKDIPNENEAQ |
| Stage | Criteria |
|---|---|
| I | Spherical, no melanin, presence of a planar clathrin coat, and few intraluminal vesicles |
| II | Oval, no melanin, presence of internal fibrils |
| III | Oval, moderate deposits of melanin onto internal fibrils |
| IV | Oval, intense deposits of melanin onto internal fibrils |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campagne, C.; Ripoll, L.; Gilles-Marsens, F.; Raposo, G.; Delevoye, C. AP-1/KIF13A Blocking Peptides Impair Melanosome Maturation and Melanin Synthesis. Int. J. Mol. Sci. 2018, 19, 568. https://doi.org/10.3390/ijms19020568
Campagne C, Ripoll L, Gilles-Marsens F, Raposo G, Delevoye C. AP-1/KIF13A Blocking Peptides Impair Melanosome Maturation and Melanin Synthesis. International Journal of Molecular Sciences. 2018; 19(2):568. https://doi.org/10.3390/ijms19020568
Chicago/Turabian StyleCampagne, Cécile, Léa Ripoll, Floriane Gilles-Marsens, Graça Raposo, and Cédric Delevoye. 2018. "AP-1/KIF13A Blocking Peptides Impair Melanosome Maturation and Melanin Synthesis" International Journal of Molecular Sciences 19, no. 2: 568. https://doi.org/10.3390/ijms19020568
APA StyleCampagne, C., Ripoll, L., Gilles-Marsens, F., Raposo, G., & Delevoye, C. (2018). AP-1/KIF13A Blocking Peptides Impair Melanosome Maturation and Melanin Synthesis. International Journal of Molecular Sciences, 19(2), 568. https://doi.org/10.3390/ijms19020568