Molecular Cloning and Characterization of Carbonic Anhydrase XII from Pufferfish (Takifugu rubripes)
Abstract
:1. Introduction
2. Results
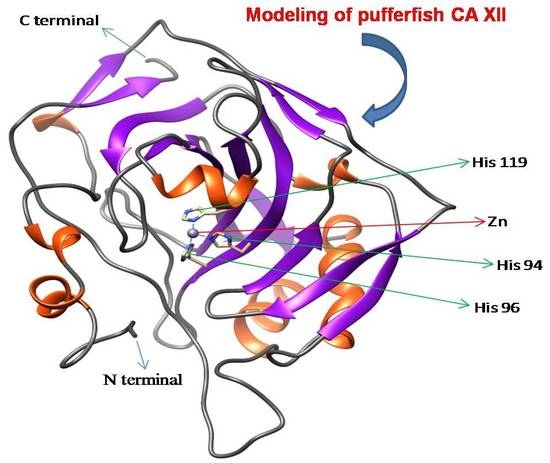
2.1. Molecular Cloning and Characterization of CA XII from Pufferfish
2.2. Expression Analysis of Pufferfish CA XII
3. Discussion
4. Materials and Methods
4.1. Experimental Fish
4.2. RNA Extraction, RT-PCR, and Molecular Cloning of CA XII
4.3. Rapid Amplification of 5′and 3′ cDNA Ends
4.4. Sequence Analysis
4.5. Phylogenetic Analysis
4.6. Template Identification and Homology Modeling of CA XII in Pufferfish
4.7. Semi-Quantitative RT-PCR Analysis of Expression
4.8. Quantitative PCR
4.9. In Situ Hybridization
4.10. Statistical Analyses
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lindskog, S.; Silverman, D. The catalytic mechanism of mammalian carbonic anhydrases. In The Carbonic Anhydrases: New Horizons; Chegwidden, W.R., Carter, N.D., Edwards, Y.H., Eds.; Birkhauser Verlag: Basel, Switzerland, 2000; pp. 175–195. [Google Scholar]
- Alterio, V.; Di Fiore, A.; D’Ambrosio, K.; Supuran, C.T.; De Simone, G. X-ray Crystallography of Carbonic Anhydrase Inhibitors and Its Importance in Drug Design; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2009; pp. 73–138. [Google Scholar]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrase inhibitors and activators for novel therapeutic applications. Future Med. Chem. 2011, 3, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Di Fiore, A.; D’Ambrosio, K.; Supuran, C.T.; De Simone, G. Multiple binding modes of inhibitors to carbonic anhydrases: How to design specific drugs targeting 15 different isoforms? Chem. Rev. 2012, 112, 4421–4468. [Google Scholar] [CrossRef] [PubMed]
- Aspatwar, A.; Tolvanen, M.E.E.; Ortutay, C.; Parkkila, S. Carbonic anhydrase related protein VIII and its role in neurodegeneration and cancer. Curr. Pharm. Des. 2010, 16, 3264–3276. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.V.; Kuzmin, I.; Wei, M.H.; Pack, S.; Geil, L.; Johnson, B.E.; Stanbridge, E.J.; Lerman, M.I. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc. Natl. Acad. Sci. USA 1998, 95, 12596–12601. [Google Scholar] [CrossRef] [PubMed]
- Türeci, Ö.; Sahin, U.; Vollmar, E.; Siemer, S.; Göttert, E.; Seitz, G.; Parkkila, A.K.; Shan, G.N.; Grubb, J.H.; Pereundschuh, M.; et al. Human carbonic anhydrase XII: cDNA cloning, expression, and chromosomal localization of a carbonic anhydrase gene that is overexpressed in some renal cell cancers. Proc. Natl. Acad. Sci. USA 1998, 95, 7608–7613. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Waheed, A.; Ulmasov, B.; Shah, G.N.; Grubb, J.H.; Sly, W.S.; Christianson, D.W. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9545–9550. [Google Scholar] [CrossRef] [PubMed]
- Kivelä, A.; Parkkila, S.; Saarnio, J.; Karttunen, T.J.; Kivelä, J.; Parkkila, A.K.; Waheed, A.; Shy, W.S.; Grubb, J.H.; Shah, G.; et al. Expression of a novel transmembrane carbonic anhydrase XII in normal human gut and colorectal tumours. Am. J. Pathol. 2000, 156, 577–584. [Google Scholar] [CrossRef]
- Halmi, P.M. Expression of Membrane-Bound Carbonic Anhydrase XII in Mouse and Rat Tissues. Master’s Thesis, University of Tempara, Tampere, Finland, 2006; p. 93. [Google Scholar]
- Tashian, R.E.; Hewett-Emmett, D.; Carter, N.; Bergenhem, N.C. Carbonic anhydrase (CA)-related proteins (CA-RPs), and transmembrane proteins with CA or CA-RP domains. In The Carbonic Anhydrases; Birkhäuser: Basel, Switzerland, 2000; pp. 105–120. [Google Scholar]
- Petrou, A.; Geronikaki, A.; Terzi, E.; Guler, O.O.; Tuccinardi, T.; Supuran, C.T. Inhibition of carbonic anhydrase isoforms I, II, IX and XII with secondary sulfonamides incorporating benzothiazole scaffolds. J. Enzyme Inhib. Med. Chem. 2016, 31, 1306–1311. [Google Scholar] [CrossRef] [PubMed]
- Fernley, R.T.; Coghlan, J.P.; Wright, R.D. Purification and characterization of a high-Mr carbonic anhydrase from sheep parotid gland. Biochem. J. 1988, 249, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Fujikawa-Adachi, K.; Nishimori, I.; Taguchi, T.; Onishi, S. Human carbonic anhydrase XIV (CA14): cDNA cloning, mRNA expression, and mapping to chromosome 1. Genomics 1999, 61, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Mori, K.; Ogawa, Y.; Ebihara, K.; Tamura, N.; Tashiro, K.; Kuwahara, T.; Mukoyama, M.; Sugawara, A.; Ozaki, S.; Tanaka, I.; et al. Isolation and characterization of CA XIV, a novel membrane-bound carbonic anhydrase from mouse kidney. J. Biol. Chem. 1999, 274, 15701–15705. [Google Scholar] [CrossRef] [PubMed]
- Opavsky, R.; Pastoreková, S.; Zelník, V.; Gibadulinová, A.; Stanbridge, E.J.; Závada, J.; Kettmann, R.; Pastorek, J. Human MN/CA9 gene, a novel member of the carbonic anhydrase family: Structure and exon to protein domain relationships. Genomics 1996, 33, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, G.J.; Kittelberger, A.M.; Watkins, R.H.; O’Reilly, M.A. Carbonic anhydrase XII mRNA encodes a hydratase that is differentially expressed along the rabbit nephron. Am. J. Physiol. 2001, 284, F399–F410. [Google Scholar] [CrossRef] [PubMed]
- Hilvo, M.; Tolvanen, M.; Clark, A.; Shen, B.; Shah, G.N.; Waheed, A.; Halmi, P.; Hänninen, M.; Hämäläinen, J.M.; Vihinen, M.; et al. Characterization of CA XV, a new GPI-anchored form of carbonic anhydrase. Biochem. J. 2005, 392, 83–92. [Google Scholar] [CrossRef] [PubMed]
- Waheed, A.; Zhu, X.L.; Sly, W.S. Membrane-associated carbonic anhydrase from rat lung. Purification, characterization, tissue distribution, and comparison with carbonic anhydrase IVs of other mammals. J. Biol. Chem. 1992, 267, 3308–3311. [Google Scholar] [PubMed]
- Zhu, X.L.; Sly, W.S. Carbonic anhydrase IV from human lung. Purification, characterization, and comparison with membrane carbonic anhydrase from human kidney. J. Biol. Chem. 1990, 265, 8795–8801. [Google Scholar] [PubMed]
- Esbaugh, A.J.; Tufts, B.L. The structure and function of carbonic anhydrase isozymes in the respiratory system of vertebrates. Respir. Physiol. Neurobiol. 2006, 154, 185–198. [Google Scholar] [CrossRef] [PubMed]
- Sumi, K.R.; Nou, I.S.; Kho, K.H. Identification and expression of a novel carbonic anhydrase isozyme in the pufferfish Takifugu vermicularis. Gene 2016, 588, 173–179. [Google Scholar] [CrossRef] [PubMed]
- Karhumaa, P.; Parkkila, S.; Türeci, O.; Waheed, A.; Grubb, J.H.; Shah, G.; Parkkila, A.; Kaunisto, K.; Tapanainen, J.; Sly, W.S.; et al. Identification of carbonic anhydrase XII as the membrane isozyme expressed in the normal human endometrial epithelium. Mol. Hum. Reprod. 2000, 6, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Parkkila, S.; Parkkila, A.K.; Saarnio, J.; Kivelä, J.; Karttunen, T.J.; Kaunisto, K.; Waheed, A.; Sly, W.S.; Türeci, O.; Virtanen, I.; et al. Expression of the membrane-associated carbonic anhydrase isozyme XII in the human kidney and renal tumors. J. Histochem. Cytochem. 2000, 48, 1601–1608. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Devi, Y. Histopathological alterations in the brain (optic tectum) of the fresh water teleost Channapunctatus in response to acute and subchronic exposure to the pesticide Chlorpyrifos. Acta Histochem. 2014, 116, 176–181. [Google Scholar] [CrossRef] [PubMed]
- Trachtenberg, M.C.; Sapirstein, V.S. Carbonic anhydrase distributions in central and peripheral nervous system of the rat. Neurochem. Res. 1980, 5, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, S.; Liao, S.Y.; Ivanova, A.; Danilkovitch-Miagkova, A.; Tarasova, N.; Weirich, G.; Merrill, M.J.; Proescholdt, M.A.; Oldfield, E.H.; Lee, J.; et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am. J. Pathol. 2001, 158, 905–919. [Google Scholar] [CrossRef]
- Wykoff, C.C.; Pugh, C.W.; Harris, A.L.; Maxwell, P.H.; Ratcliffe, P.J. The HIF pathway: Implications for patterns of gene expression in cancer. Novartis Found. Symp. 2001, 240, 212–225. [Google Scholar] [PubMed]
- Watson, P.H.; Chia, S.K.; Wykoff, C.C.; Han, C.; Leek, R.D.; Sly, W.S.; Gatter, K.C.; Ratcliffe, P.; Harris, A.L. Carbonic anhydrase XII is a marker of good prognosis in invasive breast carcinoma. Br. J. Cancer 2003, 88, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Forster, R.P.; Goldstein, L.; Rosen, K.J. Intrarenal control of urea reabsorption by renal tubules of the marine elasmobranch, Squalus acanthias. Comp. Biochem. Physiol. 1972, 42A, 2–12. [Google Scholar] [CrossRef]
- Fariselli, P.; Riccobelli, P.; Casadio, R. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins Struct. Funct. Bioinform. 1999, 36, 340–346. [Google Scholar] [CrossRef]
- Alva, V.; Nam, S.Z.; Söding, J.; Lupas, A.N. The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res. 2016, 44, W410–W415. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, D.; Lüthy, R.; Bowie, J.U. VERIFY3D: Assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997, 277, 396–404. [Google Scholar] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


 : putative active site pockets; z: zinc binding site; +: proton shuttling site; ~: substrate associated pocket; *: Thr-199 loop site. The red colored box represents the potential carbonic anhydrase domain of pufferfish CA XII. The pufferfish CA XII cloned in the present study is highlighted in bold font. GenBank accession numbers for the sequences are as follows: CA IX (human, NP_001207.2); CA XII (human, NP_996808.1; large yellow croaker, KKF18125.1; tilapia, XP_005470754.1); CA XII-like sequence (Asian arowana, KPP78156.1).
: putative active site pockets; z: zinc binding site; +: proton shuttling site; ~: substrate associated pocket; *: Thr-199 loop site. The red colored box represents the potential carbonic anhydrase domain of pufferfish CA XII. The pufferfish CA XII cloned in the present study is highlighted in bold font. GenBank accession numbers for the sequences are as follows: CA IX (human, NP_001207.2); CA XII (human, NP_996808.1; large yellow croaker, KKF18125.1; tilapia, XP_005470754.1); CA XII-like sequence (Asian arowana, KPP78156.1).
 : putative active site pockets; z: zinc binding site; +: proton shuttling site; ~: substrate associated pocket; *: Thr-199 loop site. The red colored box represents the potential carbonic anhydrase domain of pufferfish CA XII. The pufferfish CA XII cloned in the present study is highlighted in bold font. GenBank accession numbers for the sequences are as follows: CA IX (human, NP_001207.2); CA XII (human, NP_996808.1; large yellow croaker, KKF18125.1; tilapia, XP_005470754.1); CA XII-like sequence (Asian arowana, KPP78156.1).
: putative active site pockets; z: zinc binding site; +: proton shuttling site; ~: substrate associated pocket; *: Thr-199 loop site. The red colored box represents the potential carbonic anhydrase domain of pufferfish CA XII. The pufferfish CA XII cloned in the present study is highlighted in bold font. GenBank accession numbers for the sequences are as follows: CA IX (human, NP_001207.2); CA XII (human, NP_996808.1; large yellow croaker, KKF18125.1; tilapia, XP_005470754.1); CA XII-like sequence (Asian arowana, KPP78156.1).




© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumi, K.R.; Kim, S.C.; Howlader, J.; Lee, W.K.; Choi, K.S.; Kim, H.-T.; Park, J.-I.; Nou, I.-S.; Kho, K.H. Molecular Cloning and Characterization of Carbonic Anhydrase XII from Pufferfish (Takifugu rubripes). Int. J. Mol. Sci. 2018, 19, 842. https://doi.org/10.3390/ijms19030842
Sumi KR, Kim SC, Howlader J, Lee WK, Choi KS, Kim H-T, Park J-I, Nou I-S, Kho KH. Molecular Cloning and Characterization of Carbonic Anhydrase XII from Pufferfish (Takifugu rubripes). International Journal of Molecular Sciences. 2018; 19(3):842. https://doi.org/10.3390/ijms19030842
Chicago/Turabian StyleSumi, Kanij Rukshana, Soo Cheol Kim, Jewel Howlader, Won Kyo Lee, Kap Seong Choi, Hoy-Taek Kim, Jong-In Park, Ill-Sup Nou, and Kang Hee Kho. 2018. "Molecular Cloning and Characterization of Carbonic Anhydrase XII from Pufferfish (Takifugu rubripes)" International Journal of Molecular Sciences 19, no. 3: 842. https://doi.org/10.3390/ijms19030842
APA StyleSumi, K. R., Kim, S. C., Howlader, J., Lee, W. K., Choi, K. S., Kim, H. -T., Park, J. -I., Nou, I. -S., & Kho, K. H. (2018). Molecular Cloning and Characterization of Carbonic Anhydrase XII from Pufferfish (Takifugu rubripes). International Journal of Molecular Sciences, 19(3), 842. https://doi.org/10.3390/ijms19030842